to be published
Chitnumsub, P., Jaruwat, A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine hydroxymethyltransferase | 442 | Plasmodium vivax | Mutation(s): 0 Gene Names: PVC01_140059500, PVP01_1453700, PVT01_140059000 EC: 2.1.2.1 | 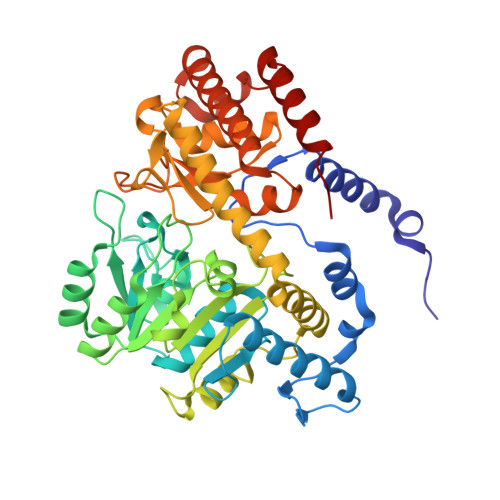 | |
UniProt | |||||
Find proteins for A5K8L9 (Plasmodium vivax (strain Salvador I)) Explore A5K8L9 Go to UniProtKB: A5K8L9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5K8L9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 8UR Query on 8UR | E [auth A], H [auth B], J [auth C] | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]piperidin-4-yl]ethanoic acid C25 H28 F3 N5 O3 UHPFIFHQJWKZHI-DEOSSOPVSA-N |  | ||
| PLG Query on PLG | D [auth A], G [auth B], I [auth C] | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] C10 H15 N2 O7 P FEVQWBMNLWUBTF-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A], K [auth C] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 101.561 | α = 90 |
| b = 58.615 | β = 90.02 |
| c = 233.052 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| PHASER | phasing |