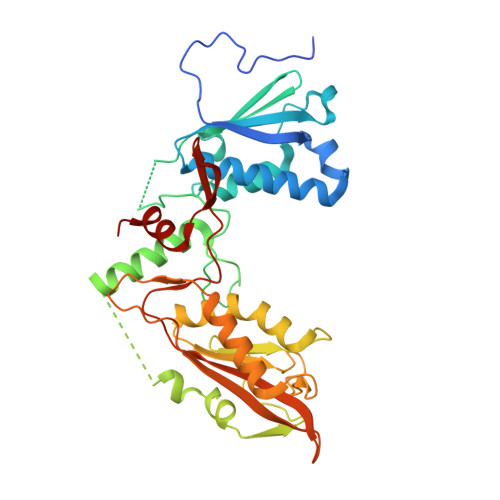
Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control
Yang, J.Y., Deng, X.Y., Li, Y.S., Ma, X.C., Feng, J.X., Yu, B., Chen, Y., Luo, Y.L., Wang, X., Chen, M.L., Fang, Z.X., Zheng, F.X., Li, Y.P., Zhong, Q., Kang, T.B., Song, L.B., Xu, R.H., Zeng, M.S., Chen, W., Zhang, H., Xie, W., Gao, S.(2018) Nat Commun 9: 1165-1165
- PubMed: 29563550
- DOI: https://doi.org/10.1038/s41467-018-03544-x
- Primary Citation of Related Structures:
5YD0 - PubMed Abstract:
Cleavage of transfer (t)RNA and ribosomal (r)RNA are critical and conserved steps of translational control for cells to overcome varied environmental stresses. However, enzymes that are responsible for this event have not been fully identified in high eukaryotes. Here, we report a mammalian tRNA/rRNA-targeting endoribonuclease: SLFN13, a member of the Schlafen family. Structural study reveals a unique pseudo-dimeric U-pillow-shaped architecture of the SLFN13 N'-domain that may clamp base-paired RNAs. SLFN13 is able to digest tRNAs and rRNAs in vitro, and the endonucleolytic cleavage dissevers 11 nucleotides from the 3'-terminus of tRNA at the acceptor stem. The cytoplasmically localised SLFN13 inhibits protein synthesis in 293T cells. Moreover, SLFN13 restricts HIV replication in a nucleolytic activity-dependent manner. According to these observations, we term SLFN13 RNase S13. Our study provides insights into the modulation of translational machinery in high eukaryotes, and sheds light on the functional mechanisms of the Schlafen family.
Organizational Affiliation:
State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-Sen University Cancer Center, Guangzhou, 510060, Guangdong, China.