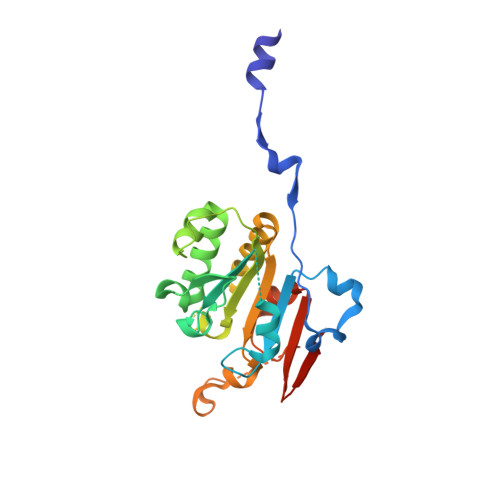
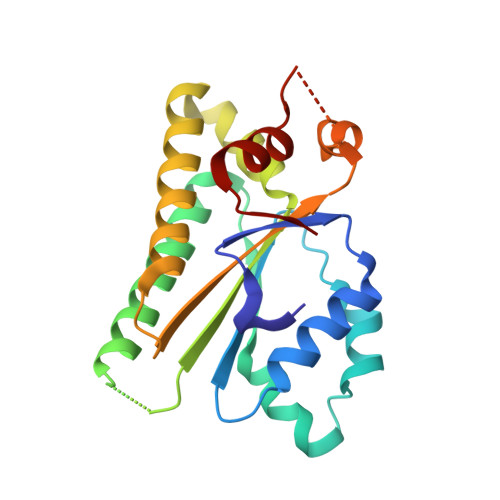
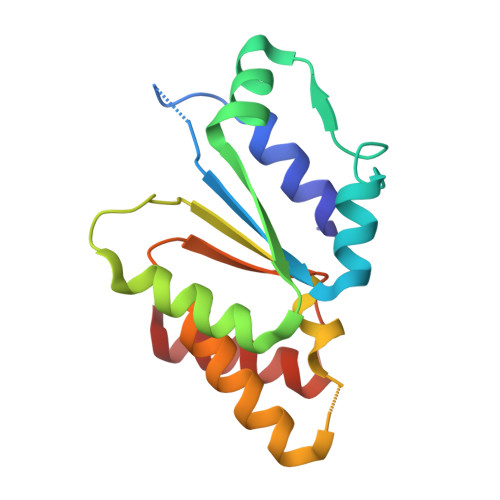
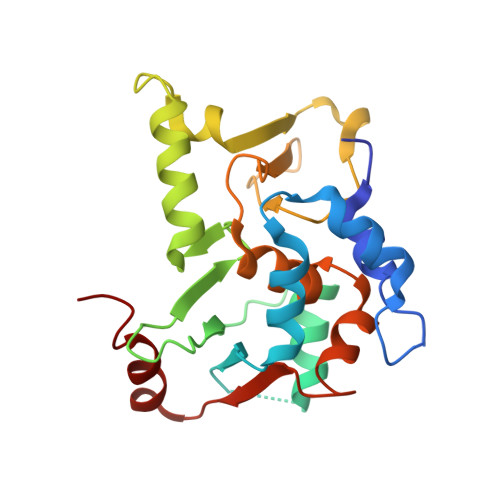
Structural basis for the functional role of the Shu complex in homologous recombination.
Zhang, S., Wang, L., Tao, Y., Bai, T., Lu, R., Zhang, T., Chen, J., Ding, J.(2017) Nucleic Acids Res 45: 13068-13079
- PubMed: 29069504
- DOI: https://doi.org/10.1093/nar/gkx992
- Primary Citation of Related Structures:
5XYN - PubMed Abstract:
The Shu complex, a conserved regulator consisting of Csm2, Psy3, Shu1 and Shu2 in budding yeast, plays an important role in the assembly of the Rad51-ssDNA filament in homologous recombination. However, the molecular basis for the assembly of the Shu complex and its functional role in DNA repair is still elusive. Here, we report the crystal structure of the yeast Shu complex, revealing that Csm2, Psy3, Shu1 and Shu2 interact with each other in sequence to form a V-shape overall structure. Shu1 adopts a structure resembling the ATPase core domain of Rad51 and represents a new Rad51 paralog. Shu2 assumes a novel structural fold consisting of a conserved zinc-finger containing SWIM domain and a small insertion domain. The functional roles of the key residues are validated using mutagenesis and in vitro pull-down and in vivo yeast growth studies. Structural analysis together with available biological data identifies two potential DNA-binding sites, one of which might be responsible for binding the ssDNA region of the 3'-overhang DNA and the other for the dsDNA region. Collectively, these findings reveal the molecular basis for the assembly of the Shu complex and shed new insight on its functional role in homologous recombination.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Science Research Center, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences; 333 Haike Road, Shanghai 201210, China.