Crystal structure of human WBSCR16, an RCC1-like protein in mitochondria
Koyama, M., Sasaki, T., Sasaki, N., Matsuura, Y.(2017) Protein Sci 26: 1870-1877
- PubMed: 28608466
- DOI: https://doi.org/10.1002/pro.3210
- Primary Citation of Related Structures:
5XGS - PubMed Abstract:
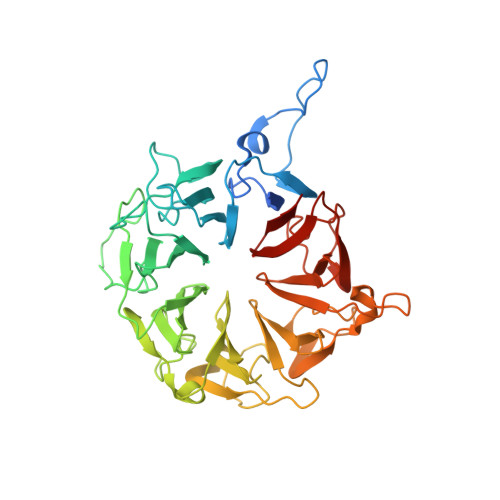
WBSCR16 (Williams-Beuren Syndrome Chromosomal Region 16) gene is located in a large deletion region of Williams-Beuren syndrome (WBS), which is a neurodevelopmental disorder. Although the relationship between WBSCR16 and WBS remains unclear, it has been reported that WBSCR16 is a member of a functional module that regulates mitochondrial 16S rRNA abundance and intra-mitochondrial translation. WBSCR16 has RCC1 (Regulator of Chromosome Condensation 1)-like amino acid sequence repeats but the function of WBSCR16 appears to be different from that of other RCC1 superfamily members. Here, we demonstrate that WBSCR16 localizes to mitochondria in HeLa cells, and report the crystal structure of WBSCR16 determined to 2.0 Å resolution using multi-wavelength anomalous diffraction. WBSCR16 adopts the seven-bladed β-propeller fold characteristic of RCC1-like proteins. A comparison of the WBSCR16 structure with that of RCC1 and other RCC1-like proteins reveals that, although many of the residues buried in the core of the β-propeller are highly conserved, the surface residues are poorly conserved and conformationally divergent.
Organizational Affiliation:
Division of Biological Science, Nagoya University, Furo-cho, Chikusa-ku, Japan.