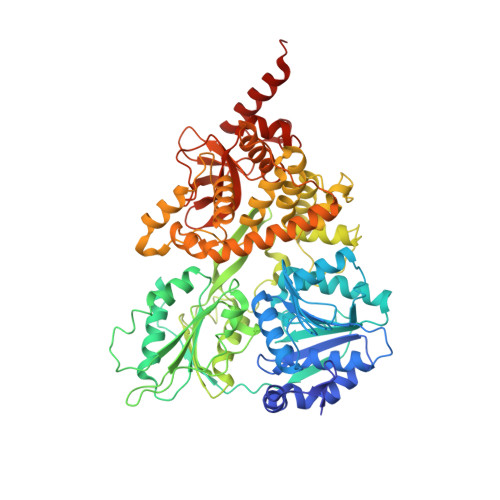
The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family
Murakami, K., Nakano, K., Shimizu, T., Ohto, U.(2017) Acta Crystallogr F Struct Biol Commun 73: 347-355
- PubMed: 28580923
- DOI: https://doi.org/10.1107/S2053230X17007336
- Primary Citation of Related Structures:
5XDR - PubMed Abstract:
DEAH-box RNA helicase 15 (DHX15) plays important roles in RNA metabolism, including in splicing and in ribosome biogenesis. In addition, mammalian DHX15 also mediates the innate immune sensing of viral RNA. However, structural information on this protein is not available, although the structure of the fungal orthologue of this protein, Prp43, has been elucidated. Here, the crystal structure of the ADP-bound form of human DHX15 is reported at a resolution of 2.0 Å. This is the first structure to be revealed of a member of the mammalian DEAH-box RNA helicase (DEAH/RHA) family in a nearly complete form, including the catalytic core consisting of the two N-terminal RecA domains and the C-terminal regulatory domains (CTD). The ADP-bound form of DHX15 displayed a compact structure, in which the RecA domains made extensive contacts with the CTD. Notably, a potential RNA-binding site was found on the surface of a RecA domain with positive electrostatic potential. Almost all structural features were conserved between the fungal Prp43 and the human DHX15, suggesting that they share a fundamentally common mechanism of action and providing a better understanding of the specific mammalian functions of DHX15.
Organizational Affiliation:
Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.