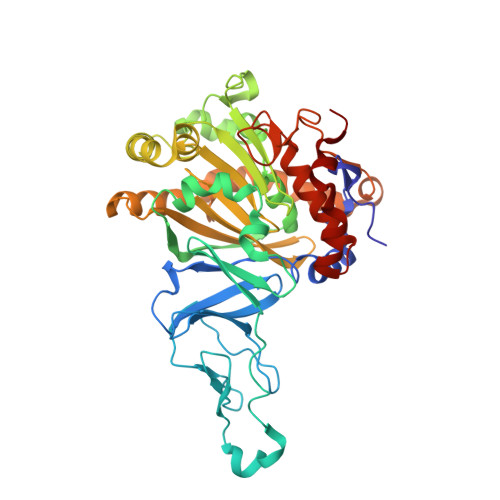
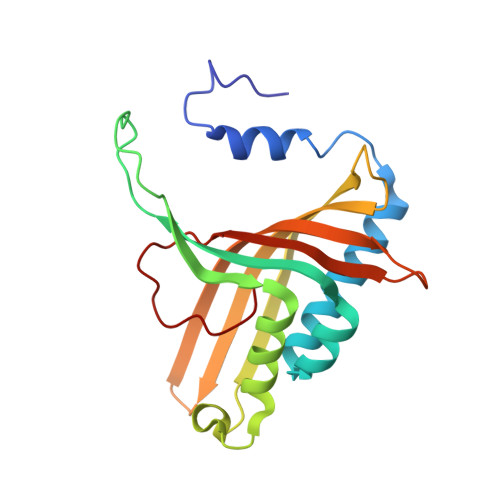
Structural and functional studies of ferredoxin and oxygenase components of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2.
Kumari, A., Singh, D., Ramaswamy, S., Ramanathan, G.(2017) PLoS One 12: e0176398-e0176398
- PubMed: 28448625
- DOI: https://doi.org/10.1371/journal.pone.0176398
- Primary Citation of Related Structures:
5XBP - PubMed Abstract:
3-nitrotoluene dioxygenase (3NTDO) from Diaphorobacter sp. strain DS2 catalyses the conversion of 3-nitrotoluene (3NT) into a mixture of 3- and 4-methylcatechols with release of nitrite. We report here, X-ray crystal structures of oxygenase and ferredoxin components of 3NTDO at 2.9 Å and 2.4 Å, respectively. The residues responsible for nitrite release in 3NTDO were further probed by four single and two double mutations in the catalytic site of α-subunit of the dioxygenase. Modification of Val 350 to Phe, Ile 204 to Ala, and Asn258 to Val by site directed mutagenesis resulted in inactive enzymes revealing the importance of these residues in catalysis. Docking studies of meta nitrotoluene to the active site of 3NTDO suggested possible orientations of binding that favor the formation of 3-methylcatechol (3MC) over 4-methylcatechol energetically. The electron transfer pathway from ferredoxin subunit to the active site of the oxygenase subunit is also proposed.
Organizational Affiliation:
Department of Chemistry, Indian Institute of Technology Kanpur, Kalyanpur, Kanpur, Uttar Pradesh, India.