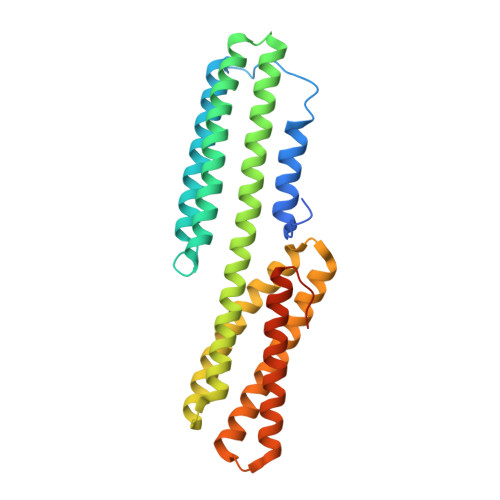
Cell-cell adhesion in metazoans relies on evolutionarily conserved features of the alpha-catenin· beta-catenin-binding interface.
Shao, X., Kang, H., Loveless, T., Lee, G.R., Seok, C., Weis, W.I., Choi, H.J., Hardin, J.(2017) J Biol Chem 292: 16477-16490
- PubMed: 28842483
- DOI: https://doi.org/10.1074/jbc.M117.795567
- Primary Citation of Related Structures:
5XA5 - PubMed Abstract:
Stable tissue integrity during embryonic development relies on the function of the cadherin·catenin complex (CCC). The Caenorhabditis elegans CCC is a useful paradigm for analyzing in vivo requirements for specific interactions among the core components of the CCC, and it provides a unique opportunity to examine evolutionarily conserved mechanisms that govern the interaction between α- and β-catenin. HMP-1, unlike its mammalian homolog α-catenin, is constitutively monomeric, and its binding affinity for HMP-2/β-catenin is higher than that of α-catenin for β-catenin. A crystal structure shows that the HMP-1·HMP-2 complex forms a five-helical bundle structure distinct from the structure of the mammalian α-catenin·β-catenin complex. Deletion analysis based on the crystal structure shows that the first helix of HMP-1 is necessary for binding HMP-2 avidly in vitro and for efficient recruitment of HMP-1 to adherens junctions in embryos. HMP-2 Ser-47 and Tyr-69 flank its binding interface with HMP-1, and we show that phosphomimetic mutations at these two sites decrease binding affinity of HMP-1 to HMP-2 by 40-100-fold in vitro. In vivo experiments using HMP-2 S47E and Y69E mutants showed that they are unable to rescue hmp-2 ( zu364 ) mutants, suggesting that phosphorylation of HMP-2 on Ser-47 and Tyr-69 could be important for regulating CCC formation in C. elegans Our data provide novel insights into how cadherin-dependent cell-cell adhesion is modulated in metazoans by conserved elements as well as features unique to specific organisms.
Organizational Affiliation:
From the Program in Genetics.