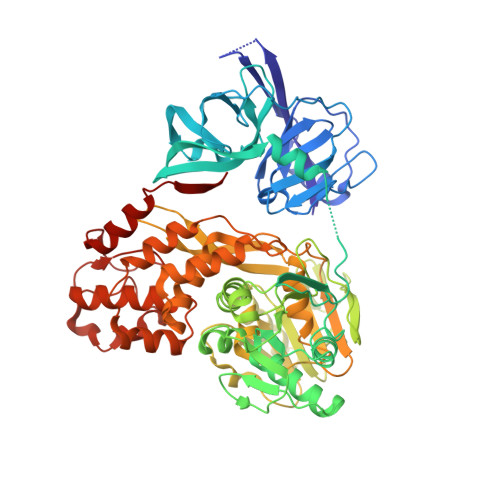
NS3 from Hepatitis C Virus Strain JFH-1 Is an Unusually Robust Helicase That Is Primed To Bind and Unwind Viral RNA.
Zhou, T., Ren, X., Adams, R.L., Pyle, A.M.(2018) J Virol 92
- PubMed: 29070684
- DOI: https://doi.org/10.1128/JVI.01253-17
- Primary Citation of Related Structures:
5WDX - PubMed Abstract:
Hepatitis C viruses (HCV) encode a helicase enzyme that is essential for viral replication and assembly (nonstructural protein 3 [NS3]). This helicase has become the focus of extensive basic research on the general helicase mechanism, and it is also of interest as a novel drug target. Despite the importance of this protein, mechanistic work on NS3 has been conducted almost exclusively on variants from HCV genotype 1. Our understanding of NS3 from the highly active HCV strains that are used to study HCV genetics and mechanism in cell culture (such as JFH-1) is lacking. We therefore set out to determine whether NS3 from the replicatively efficient genotype 2a strain JFH-1 displays novel functional or structural properties. Using biochemical assays for RNA binding and duplex unwinding, we show that JFH-1 NS3 binds RNA much more rapidly than the previously studied NS3 variants from genotype 1b. Unlike NS3 variants from other genotypes, JFH-1 NS3 binds RNA with high affinity in a functionally active form that is capable of immediately unwinding RNA duplexes without undergoing rate-limiting conformational changes that precede activation. Unlike other superfamily 2 (SF2) helicases, JFH-1 NS3 does not require long 3' overhangs, and it unwinds duplexes that are flanked by only a few nucleotides, as in the folded HCV genome. To understand the physical basis for this, we solved the crystal structure of JFH-1 NS3, revealing a novel conformation that contains an open, positively charged RNA binding cleft that is primed for productive interaction with RNA targets, potentially explaining robust replication by HCV JFH-1. IMPORTANCE Genotypes of HCV are as divergent as different types of flavivirus, and yet mechanistic features of HCV variants are presumed to be held in common. One of the most well-studied components of the HCV replication complex is a helicase known as nonstructural protein 3 (NS3). We set out to determine whether this important mechanical component possesses biochemical and structural properties that differ between common strains such as those of genotype 1b and a strain of HCV that replicates with exceptional efficiency (JFH-1, classified as genotype 2a). Indeed, unlike the inefficient genotype 1b NS3, which has been well studied, JFH-1 NS3 is a superhelicase with strong RNA affinity and high unwinding efficiency on a broad range of targets. Crystallographic analysis reveals architectural features that promote enhanced biochemical activity of JFH-1 NS3. These findings show that even within a single family of viruses, drift in sequence can result in the acquisition of radically new functional properties that enhance viral fitness.
Organizational Affiliation:
Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, Connecticut, USA.