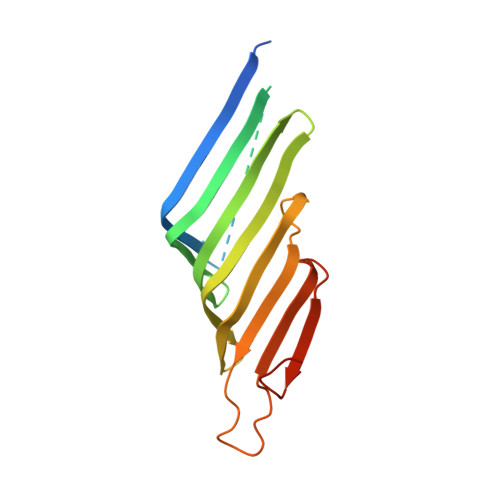
The Structure of a Conserved Domain of TamB Reveals a Hydrophobic beta Taco Fold.
Josts, I., Stubenrauch, C.J., Vadlamani, G., Mosbahi, K., Walker, D., Lithgow, T., Grinter, R.(2017) Structure 25: 1898-1906.e5
- PubMed: 29129383
- DOI: https://doi.org/10.1016/j.str.2017.10.002
- Primary Citation of Related Structures:
5VTG - PubMed Abstract:
The translocation and assembly module (TAM) plays a role in the transport and insertion of proteins into the bacterial outer membrane. TamB, a component of this system spans the periplasmic space to engage with its partner protein TamA. Despite efforts to characterize the TAM, the structure and mechanism of action of TamB remained enigmatic. Here we present the crystal structure of TamB amino acids 963-1,138. This region represents half of the conserved DUF490 domain, the defining feature of TamB. TamB 963-1138 consists of a concave, taco-shaped β sheet with a hydrophobic interior. This β taco structure is of dimensions capable of accommodating and shielding the hydrophobic side of an amphipathic β strand, potentially allowing TamB to chaperone nascent membrane proteins from the aqueous environment. In addition, sequence analysis suggests that the structure of TamB 963-1138 is shared by a large portion of TamB. This architecture could allow TamB to act as a conduit for membrane proteins.
Organizational Affiliation:
The Hamburg Centre for Ultrafast Imaging (CUI), Institute for Biochemistry and Molecular Biology, University of Hamburg, Martin-Luther-King-Platz 6, 20146 Hamburg, Germany; Department of Chemistry, Institute for Biochemistry and Molecular Biology, University of Hamburg, Martin-Luther-King-Platz 6, 20146 Hamburg, Germany.