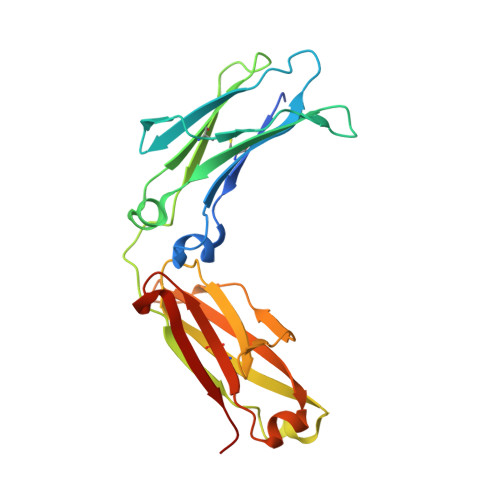
Structure of the Fc fragment of the NIST reference antibody RM8671.
Gallagher, D.T., Galvin, C.V., Karageorgos, I.(2018) Acta Crystallogr F Struct Biol Commun 74: 524-529
- PubMed: 30198883
- DOI: https://doi.org/10.1107/S2053230X18009834
- Primary Citation of Related Structures:
5VGP - PubMed Abstract:
As the link between antigen binding and immune activation, the antibody Fc region has received extensive structural study. In this report, the structure of the Fc fragment of the NIST IgG1 mAb (reference material 8671) is described at 2.1 Å resolution in space group P2 1 2 1 2 1 , with approximate unit-cell parameters a = 50, b = 80, c = 138 Å. Prior Fc structures with a wide variety of modifications are also surveyed, focusing on those in the same crystal form. To facilitate the analysis of conformations, a reference frame and a two-parameter metric are proposed, considering the CH2 domains as mobile with respect to a fixed dimeric CH3 core. Over several human Fc structures, a significant variation in Fc elbow conformations is observed, which may serve to facilitate the regulation of Fc effector signaling.
Organizational Affiliation:
NIST/IBBR, 9600 Gudelsky Drive, Rockville, MD 20850, USA.