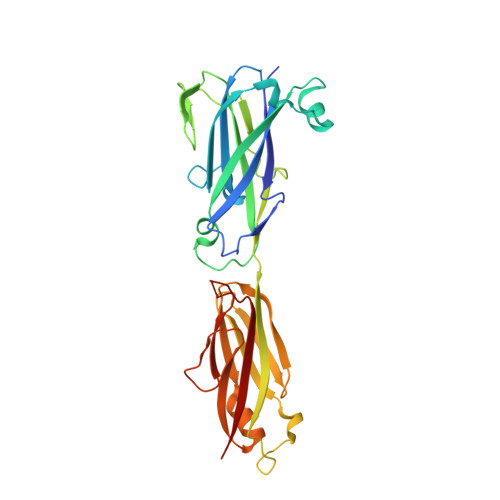
Domain structure and cross-linking in a giant adhesin from the Mobiluncus mulieris bacterium.
Young, P.G., Paynter, J.M., Wardega, J.K., Middleditch, M.J., Payne, L.S., Baker, E.N., Squire, C.J.(2023) Acta Crystallogr D Struct Biol 79: 971-979
- PubMed: 37860959
- DOI: https://doi.org/10.1107/S2059798323007507
- Primary Citation of Related Structures:
5U5O, 5U6F - PubMed Abstract:
Cell-surface proteins known as adhesins enable bacteria to colonize particular environments, and in Gram-positive bacteria often contain autocatalytically formed covalent intramolecular cross-links. While investigating the prevalence of such cross-links, a remarkable example was discovered in Mobiluncus mulieris, a pathogen associated with bacterial vaginosis. This organism encodes a putative adhesin of 7651 residues. Crystallography and mass spectrometry of two selected domains, and AlphaFold structure prediction of the remainder of the protein, were used to show that this adhesin belongs to the family of thioester, isopeptide and ester-bond-containing proteins (TIE proteins). It has an N-terminal domain homologous to thioester adhesion domains, followed by 51 immunoglobulin (Ig)-like domains containing ester- or isopeptide-bond cross-links. The energetic cost to the M. mulieris bacterium in retaining such a large adhesin as a single gene or protein construct suggests a critical role in pathogenicity and/or persistence.
Organizational Affiliation:
School of Biological Sciences, The University of Auckland, Private Bag 92019, Auckland 1010, New Zealand.