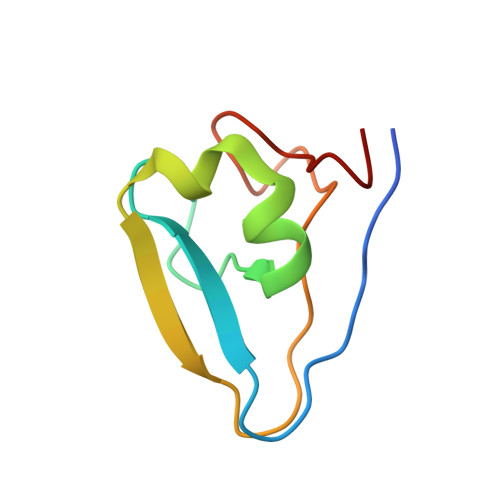
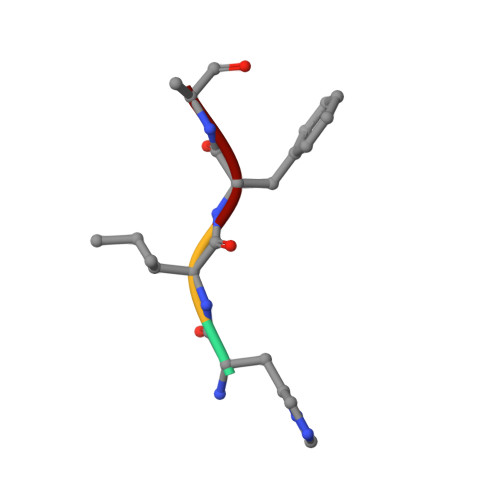
Bound Waters Mediate Binding of Diverse Substrates to a Ubiquitin Ligase.
Munoz-Escobar, J., Matta-Camacho, E., Cho, C., Kozlov, G., Gehring, K.(2017) Structure 25: 719-729.e3
- PubMed: 28392261
- DOI: https://doi.org/10.1016/j.str.2017.03.004
- Primary Citation of Related Structures:
5TDA, 5TDB, 5TDC, 5TDD, 5UM3 - PubMed Abstract:
The N-end rule pathway controls the half-life of proteins based on their N-terminal residue. Positively charged type 1 N-degrons are recognized by a negatively charged pocket on the Zn finger named the UBR box. Here, we show that the UBR box is rigid, but bound water molecules in the pocket provide the structural plasticity required to bind different positively charged amino acids. Ultra-high-resolution crystal structures of arginine, histidine, and methylated arginine reveal that water molecules mediate the binding of N-degron peptides. Using a high-throughput binding assay and isothermal titration calorimetry, we demonstrate that the UBR box is able to bind methylated arginine and lysine peptides with high affinity and measure the preference for hydrophobic residues in the second position in the N-degron peptide. Finally, we show that the V122L mutation present in Johanson-Blizzard syndrome patients changes the specificity for the second position due to occlusion of the secondary pocket.
Organizational Affiliation:
Department of Biochemistry, Groupe de Recherche Axé sur la Structure des Protéines, McGill University, Montreal, QC H3G 0B1, Canada.