The crystal structure of the Leishmania infantum Silent Information Regulator 2 related protein 1: Implications to protein function and drug design.
Ronin, C., Costa, D.M., Tavares, J., Faria, J., Ciesielski, F., Ciapetti, P., Smith, T.K., MacDougall, J., Cordeiro-da-Silva, A., Pemberton, I.K.(2018) PLoS One 13: e0193602-e0193602
- PubMed: 29543820
- DOI: https://doi.org/10.1371/journal.pone.0193602
- Primary Citation of Related Structures:
5OL0 - PubMed Abstract:
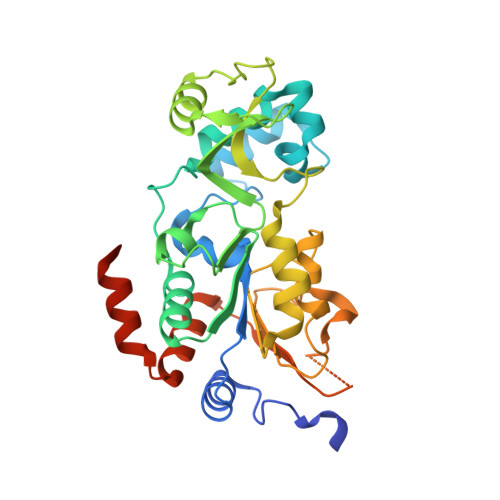
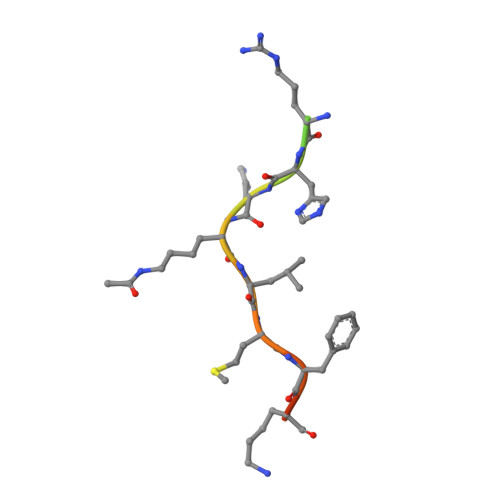
The de novo crystal structure of the Leishmania infantum Silent Information Regulator 2 related protein 1 (LiSir2rp1) has been solved at 1.99Å in complex with an acetyl-lysine peptide substrate. The structure is broadly commensurate with Hst2/SIRT2 proteins of yeast and human origin, reproducing many of the structural features common to these sirtuin deacetylases, including the characteristic small zinc-binding domain, and the larger Rossmann-fold domain involved in NAD+-binding interactions. The two domains are linked via a cofactor binding loop ordered in open conformation. The peptide substrate binds to the LiSir2rp1 protein via a cleft formed between the small and large domains, with the acetyl-lysine side chain inserting further into the resultant hydrophobic tunnel. Crystals were obtained only with recombinant LiSir2rp1 possessing an extensive internal deletion of a proteolytically-sensitive region unique to the sirtuins of kinetoplastid origin. Deletion of 51 internal amino acids (P253-E303) from LiSir2rp1 did not appear to alter peptide substrate interactions in deacetylation assays, but was indispensable to obtain crystals. Removal of this potentially flexible region, that otherwise extends from the classical structural elements of the Rossmann-fold, specifically the β8-β9 connector, appears to result in lower accumulation of the protein when expressed from episomal vectors in L. infantum SIR2rp1 single knockout promastigotes. The biological function of the large serine-rich insertion in kinetoplastid/trypanosomatid sirtuins, highlighted as a disordered region with strong potential for post-translational modification, remains unknown but may confer additional cellular functions that are distinct from their human counterparts. These unique molecular features, along with the resolution of the first kinetoplastid sirtuin deacetylase structure, present novel opportunities for drug design against a protein target previously established as essential to parasite survival and proliferation.
Organizational Affiliation:
NovAliX - Bioparc, Bd Sébastien Brant, Illkirch, France.