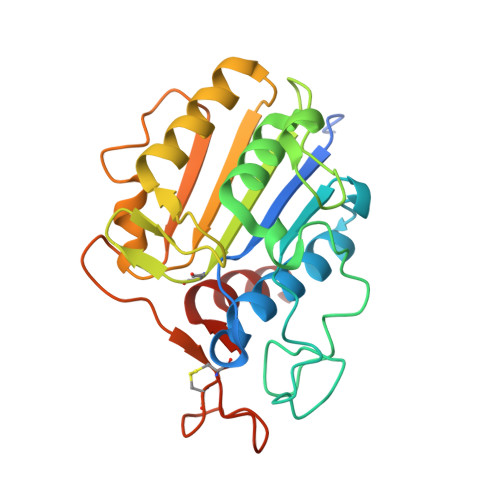
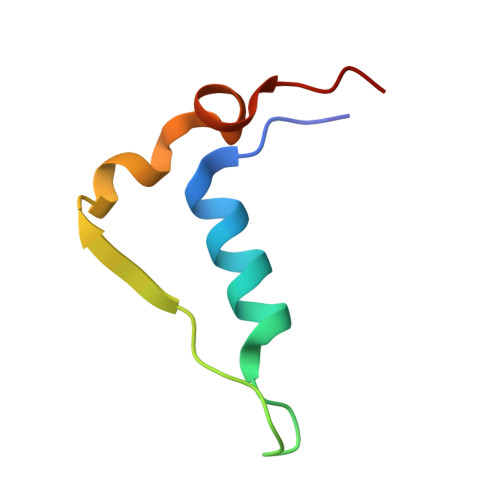
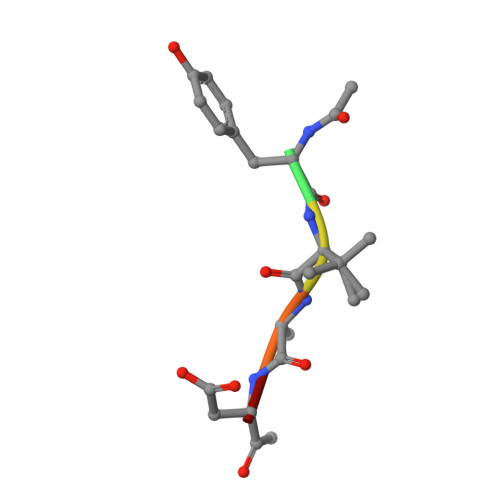
Structural analyses ofArabidopsis thalianalegumain gamma reveal differential recognition and processing of proteolysis and ligation substrates.
Zauner, F.B., Elsasser, B., Dall, E., Cabrele, C., Brandstetter, H.(2018) J Biol Chem 293: 8934-8946
- PubMed: 29628443
- DOI: https://doi.org/10.1074/jbc.M117.817031
- PubMed Abstract:
Legumain is a dual-function protease-peptide ligase whose activities are of great interest to researchers studying plant physiology and to biotechnological applications. However, the molecular mechanisms determining the specificities for proteolysis and ligation are unclear because structural information on the substrate recognition by a fully activated plant legumain is unavailable. Here, we present the X-ray structure of Arabidopsis thaliana legumain isoform γ (AtLEGγ) in complex with the covalent peptidic Ac-YVAD chloromethyl ketone (CMK) inhibitor targeting the catalytic cysteine. Mapping of the specificity pockets preceding the substrate-cleavage site explained the known substrate preference. The comparison of inhibited and free AtLEGγ structures disclosed a substrate-induced disorder-order transition with synergistic rearrangements in the substrate-recognition sites. Docking and in vitro studies with an AtLEGγ ligase substrate, sunflower trypsin inhibitor (SFTI), revealed a canonical, protease substrate-like binding to the active site-binding pockets preceding and following the cleavage site. We found the interaction of the second residue after the scissile bond, P2'-S2', to be critical for deciding on proteolysis versus cyclization. cis-trans -Isomerization of the cyclic peptide product triggered its release from the AtLEGγ active site and prevented inadvertent cleavage. The presented integrative mechanisms of proteolysis and ligation (transpeptidation) explain the interdependence of legumain and its preferred substrates and provide a rational framework for engineering optimized proteases, ligases, and substrates.
Organizational Affiliation:
From the Department of Biosciences, University of Salzburg, Salzburg 5020, Austria.