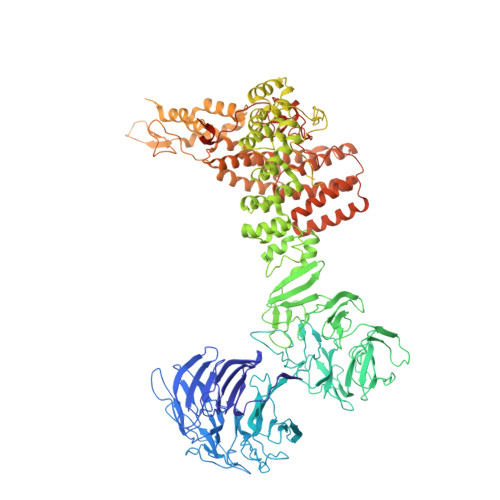
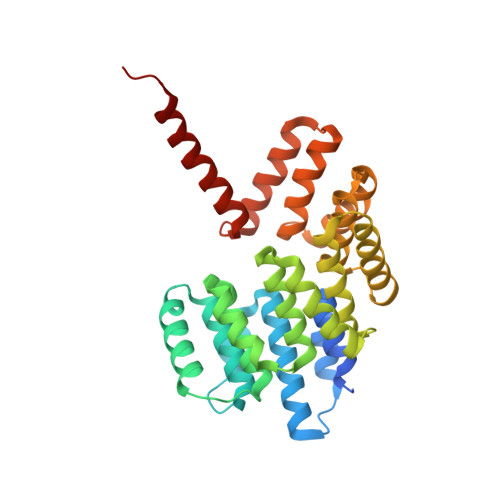
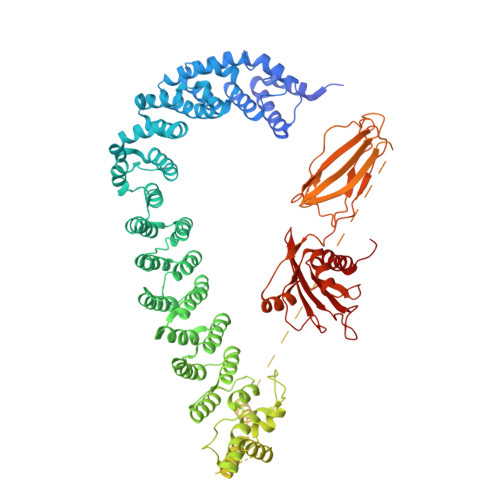
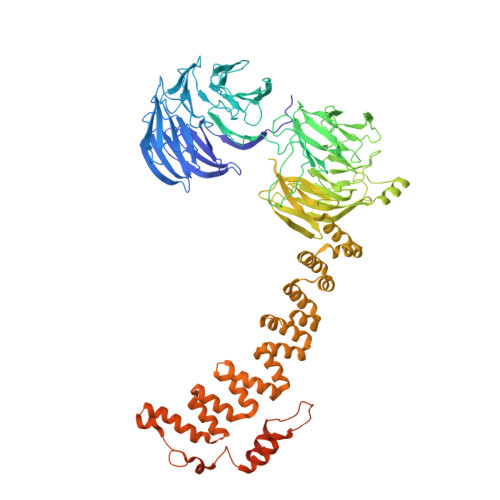
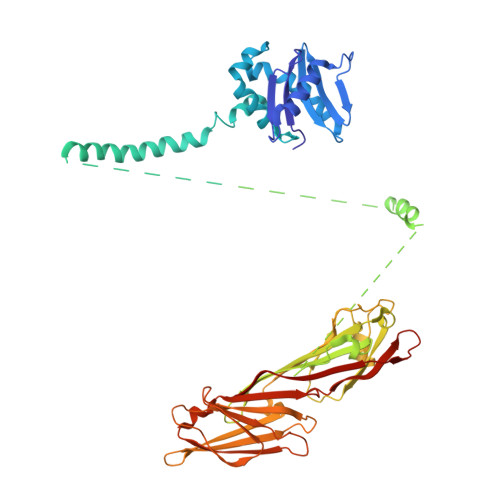
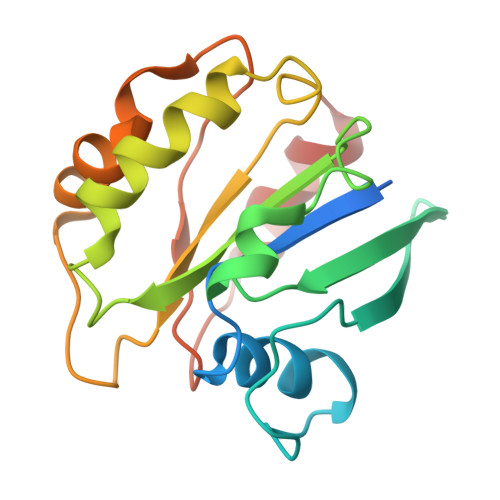
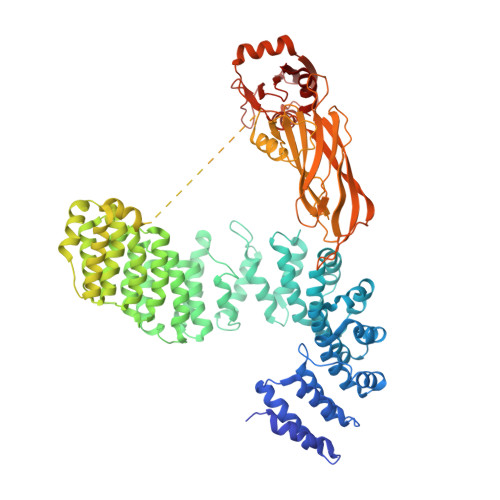
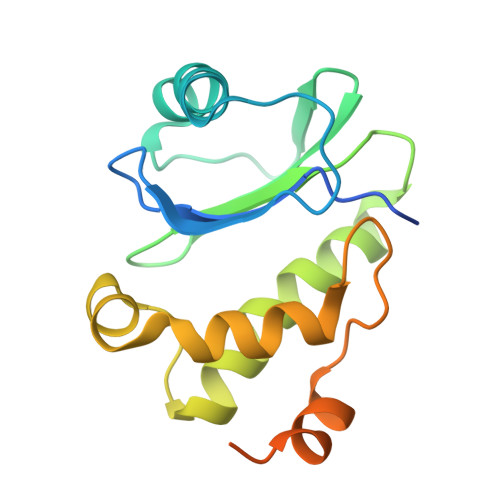
9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Dodonova, S.O., Aderhold, P., Kopp, J., Ganeva, I., Rohling, S., Hagen, W.J., Sinning, I., Wieland, F., Briggs, J.A.(2017) Elife 6
- PubMed: 28621666
- DOI: https://doi.org/10.7554/eLife.26691
- Primary Citation of Related Structures:
5MU7, 5NZR, 5NZS, 5NZT, 5NZU, 5NZV - PubMed Abstract:
COPI coated vesicles mediate trafficking within the Golgi apparatus and between the Golgi and the endoplasmic reticulum. Assembly of a COPI coated vesicle is initiated by the small GTPase Arf1 that recruits the coatomer complex to the membrane, triggering polymerization and budding. The vesicle uncoats before fusion with a target membrane. Coat components are structurally conserved between COPI and clathrin/adaptor proteins. Using cryo-electron tomography and subtomogram averaging, we determined the structure of the COPI coat assembled on membranes in vitro at 9 Å resolution. We also obtained a 2.57 Å resolution crystal structure of βδ-COP. By combining these structures we built a molecular model of the coat. We additionally determined the coat structure in the presence of ArfGAP proteins that regulate coat dissociation. We found that Arf1 occupies contrasting molecular environments within the coat, leading us to hypothesize that some Arf1 molecules may regulate vesicle assembly while others regulate coat disassembly.
Organizational Affiliation:
Structural and Computational Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany.