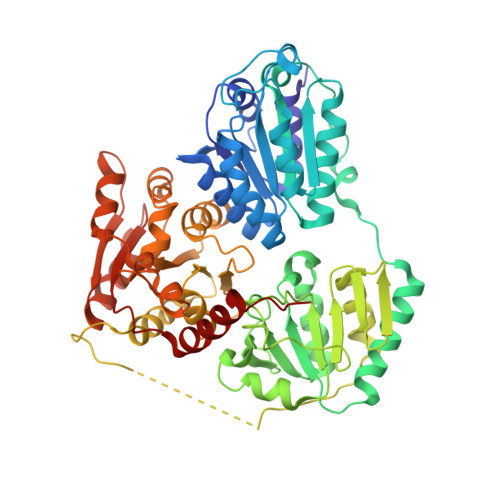
Crystal structure of an inferred ancestral bacterial pyruvate decarboxylase.
Buddrus, L., Andrews, E.S.V., Leak, D.J., Danson, M.J., Arcus, V.L., Crennell, S.J.(2018) Acta Crystallogr F Struct Biol Commun 74: 179-186
- PubMed: 29497023
- DOI: https://doi.org/10.1107/S2053230X18002819
- Primary Citation of Related Structures:
5NPU - PubMed Abstract:
Pyruvate decarboxylase (PDC; EC 4.1.1.1) is a key enzyme in homofermentative metabolism where ethanol is the major product. PDCs are thiamine pyrophosphate- and Mg 2+ ion-dependent enzymes that catalyse the non-oxidative decarboxylation of pyruvate to acetaldehyde and carbon dioxide. As this enzyme class is rare in bacteria, current knowledge of bacterial PDCs is extremely limited. One approach to further the understanding of bacterial PDCs is to exploit the diversity provided by evolution. Ancestral sequence reconstruction (ASR) is a method of computational molecular evolution to infer extinct ancestral protein sequences, which can then be synthesized and experimentally characterized. Through ASR a novel PDC was generated, designated ANC27, that shares only 78% amino-acid sequence identity with its closest extant homologue (Komagataeibacter medellinensis PDC, GenBank accession No. WP_014105323.1), yet is fully functional. Crystals of this PDC diffracted to 3.5 Å resolution. The data were merged in space group P3 2 21, with unit-cell parameters a = b = 108.33, c = 322.65 Å, and contained two dimers (two tetramer halves) in the asymmetric unit. The structure was solved by molecular replacement using PDB entry 2wvg as a model, and the final R values were R work = 0.246 (0.3671 in the highest resolution bin) and R free = 0.319 (0.4482 in the highest resolution bin). Comparison with extant bacterial PDCs supports the previously observed correlation between decreased tetramer interface area (and number of interactions) and decreased thermostability.
Organizational Affiliation:
School of Biochemistry, University of Bristol, University Walk, Bristol BS8 1TD, England.