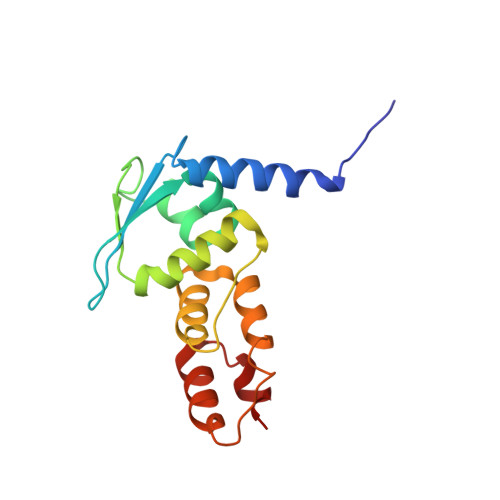
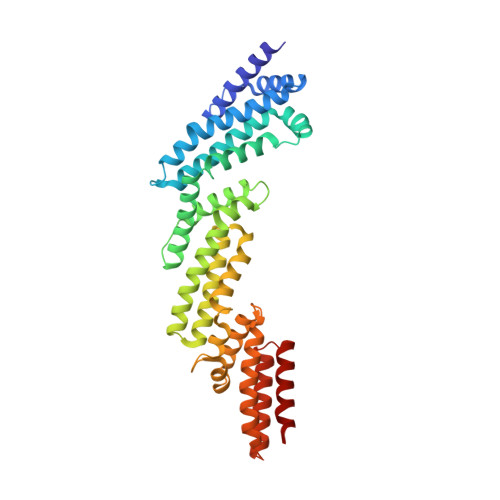
Structural and biochemical characterization establishes a detailed understanding of KEAP1-CUL3 complex assembly.
Adamson, R.J., Payne, N.C., Bartual, S.G., Mazitschek, R., Bullock, A.N.(2023) Free Radic Biol Med 204: 215-225
- PubMed: 37156295
- DOI: https://doi.org/10.1016/j.freeradbiomed.2023.04.021
- Primary Citation of Related Structures:
5NLB - PubMed Abstract:
KEAP1 promotes the ubiquitin-dependent degradation of NRF2 by assembling into a CUL3-dependent ubiquitin ligase complex. Oxidative and electrophilic stress inhibit KEAP1 allowing NRF2 to accumulate for the transactivation of stress response genes. To date there are no structures of the KEAP1-CUL3 interaction nor binding data to show the contributions of different domains to their binding affinity. We determined a crystal structure of the BTB and 3-box domains of human KEAP1 in complex with the CUL3 N-terminal domain that showed a heterotetrameric assembly with 2:2 stoichiometry. To support the structural data, we developed a versatile TR-FRET-based assay system to profile the binding of BTB-domain-containing proteins to CUL3 and determine the contribution of distinct protein features, revealing the importance of the CUL3 N-terminal extension for high affinity binding. We further provide direct evidence that the investigational drug CDDO does not disrupt the KEAP1-CUL3 interaction, even at high concentrations, but reduces the affinity of KEAP1-CUL3 binding. The TR-FRET-based assay system offers a generalizable platform for profiling this protein class and may form a suitable screening platform for ligands that disrupt these interactions by targeting the BTB or 3-box domains to block E3 ligase function.
Organizational Affiliation:
Centre for Medicines Discovery, Nuffield Department of Medicine, University of Oxford, Oxford, OX3 7FZ, UK.