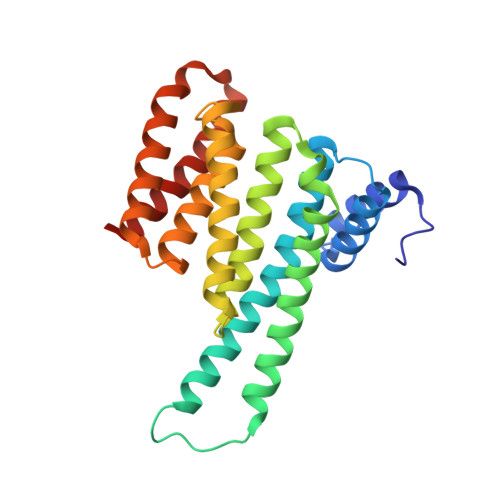
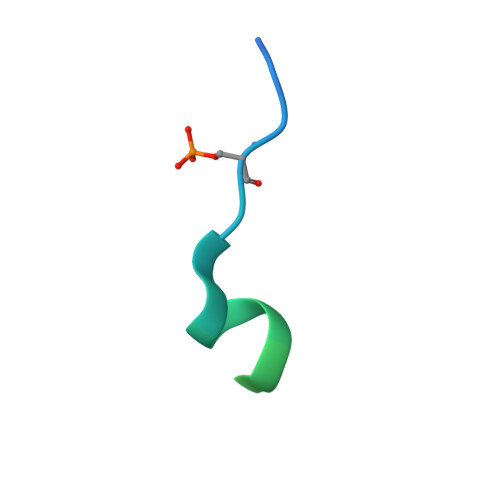
Structural interface between LRRK2 and 14-3-3 protein.
Stevers, L.M., de Vries, R.M., Doveston, R.G., Milroy, L.G., Brunsveld, L., Ottmann, C.(2017) Biochem J 474: 1273-1287
- PubMed: 28202711
- DOI: https://doi.org/10.1042/BCJ20161078
- Primary Citation of Related Structures:
5MY9, 5MYC - PubMed Abstract:
Binding of 14-3-3 proteins to leucine-rich repeat protein kinase 2 (LRRK2) is known to be impaired by many Parkinson's disease (PD)-relevant mutations. Abrogation of this interaction is connected to enhanced LRRK2 kinase activity, which in turn is implicated in increased ubiquitination of LRRK2, accumulation of LRRK2 into inclusion bodies and reduction in neurite length. Hence, the interaction between 14-3-3 and LRRK2 is of significant interest as a possible drug target for the treatment of PD. However, LRRK2 possesses multiple sites that, upon phosphorylation, can bind to 14-3-3, thus rendering the interaction relatively complex. Using biochemical assays and crystal structures, we characterize the multivalent interaction between these two proteins.
Organizational Affiliation:
Laboratory of Chemical Biology, Department of Biomedical Engineering and Institute for Complex Molecular Systems, Eindhoven University of Technology, PO Box 513, 5600 MB Eindhoven, The Netherlands.