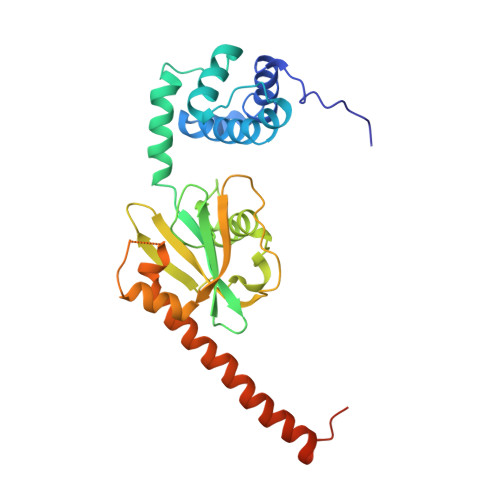
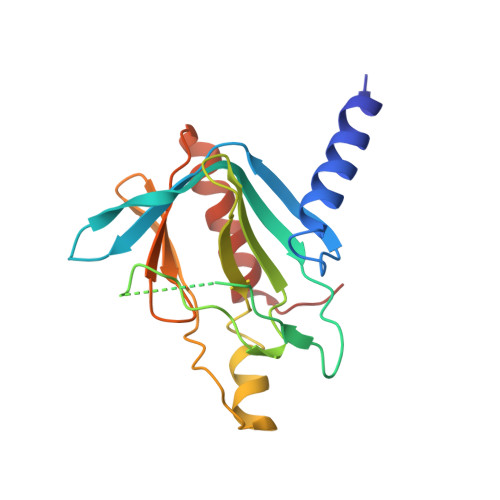
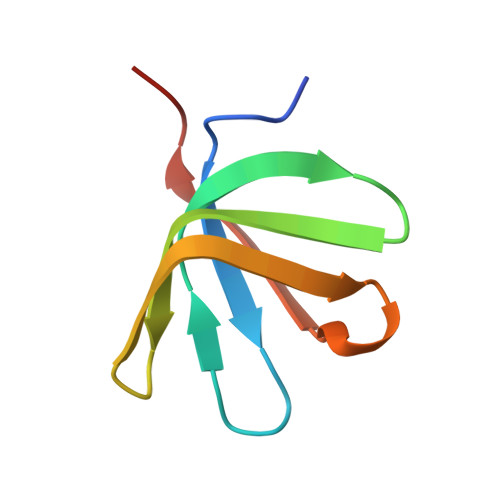
Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Charenton, C., Taverniti, V., Gaudon-Plesse, C., Back, R., Seraphin, B., Graille, M.(2016) Nat Struct Mol Biol 23: 982-986
- PubMed: 27694841
- DOI: https://doi.org/10.1038/nsmb.3300
- Primary Citation of Related Structures:
5LON, 5LOP - PubMed Abstract:
Elimination of the 5' cap of eukaryotic mRNAs, known as decapping, is considered to be a crucial, irreversible and highly regulated step required for the rapid degradation of mRNA by Xrn1, the major cytoplasmic 5'-3' exonuclease. Decapping is accomplished by the recruitment of a protein complex formed by the Dcp2 catalytic subunit and its Dcp1 cofactor. However, this complex has a low intrinsic enzymatic activity and requires several accessory proteins such as the Lsm1-7 complex, Pat1, Edc1-Edc2 and/or Edc3 to be fully active. Here we present the crystal structure of the active form of the yeast Kluyveromyces lactis Dcp1-Dcp2 enzyme bound to its product (m 7 GDP) and its potent activator Edc3. This structure of the Dcp1-Dcp2 complex bound to a cap analog further explains previously published data on substrate binding and provides hints as to the mechanism of Edc3-mediated Dcp2 activation.
Organizational Affiliation:
Laboratoire de Biochimie, Ecole Polytechnique, CNRS, Université Paris-Saclay, Palaiseau, France.