Differing self-assembling protein nanostructures in soluble and crystal form reveal dynamic behaviour
Cronin, S.L., Navon, Y., Zarivach, R., Bitton, R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Magnetosome protein MamA | 202 | Solidesulfovibrio magneticus RS-1 | Mutation(s): 0 Gene Names: mamA, DMR_41160 | 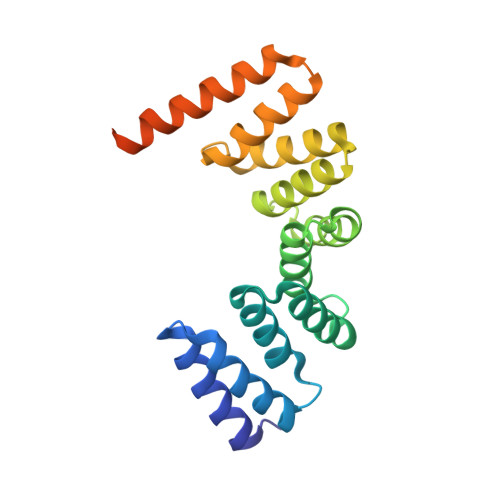 | |
UniProt | |||||
Find proteins for C4XPQ7 (Solidesulfovibrio magneticus (strain ATCC 700980 / DSM 13731 / RS-1)) Explore C4XPQ7 Go to UniProtKB: C4XPQ7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C4XPQ7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL Query on CL | G [auth A] H [auth B] I [auth C] J [auth C] K [auth D] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 150.548 | α = 90 |
| b = 150.548 | β = 90 |
| c = 204.297 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| BALBES | phasing |