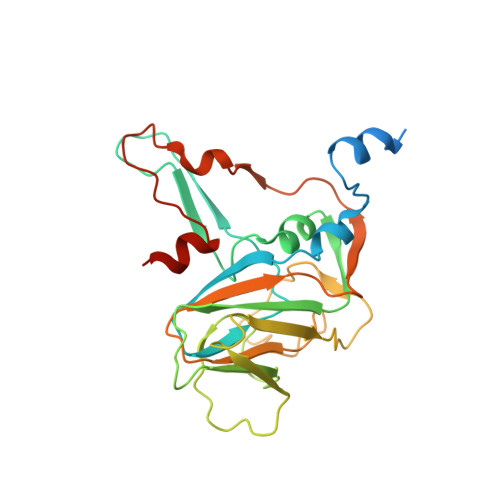
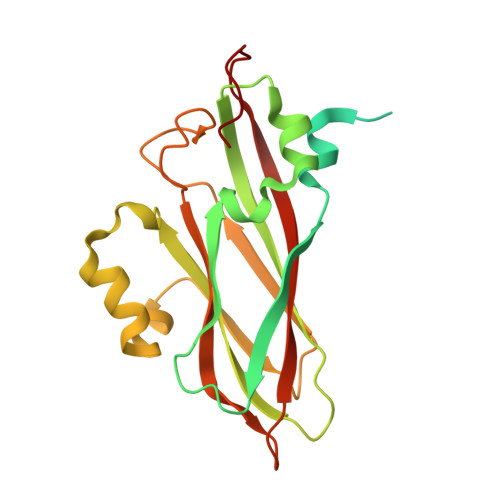
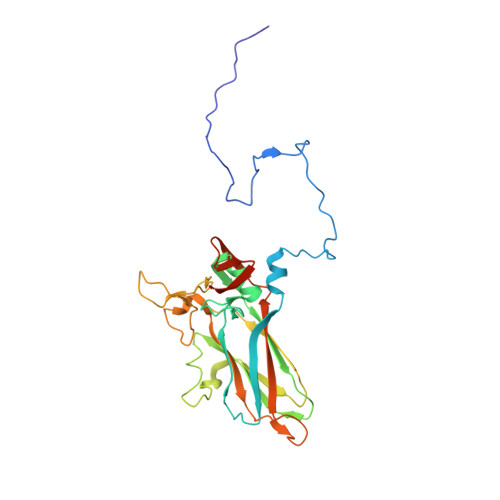
X-ray structure of Triatoma virus empty capsid: insights into the mechanism of uncoating and RNA release in dicistroviruses.
Sanchez-Eugenia, R., Durana, A., Lopez-Marijuan, I., Marti, G.A., Guerin, D.M.(2016) J Gen Virol 97: 2769-2779
- PubMed: 27519423
- DOI: https://doi.org/10.1099/jgv.0.000580
- Primary Citation of Related Structures:
5L7O - PubMed Abstract:
In viruses, uncoating and RNA release are two key steps of successfully infecting a target cell. During these steps, the capsid must undergo the necessary conformational changes to allow RNA egress. Despite their importance, these processes are poorly understood in the family Dicistroviridae. Here, we used X-ray crystallography to solve the atomic structure of a Triatoma virus(TrV) empty particle (Protein Data Bank ID 5L7O), which is the resulting capsid after RNA release. It is observed that the overall shape of the capsid and of the three individual proteins is maintained in comparison with the mature virion. Furthermore, no channels indicative of RNA release are formed in the TrV empty particle. However, the most prominent change in the empty particle when compared with the mature virion is the loss of order in the N-terminal domain of the VP2 protein. In mature virions, the VP2 N-terminal domain of one pentamer is swapped with its twofold related copy in an adjacent pentamer, thereby stabilizing the binding between the pentamers. The loss of these interactions allows us to propose that RNA release may take place through transient flipping-out of pentameric subunits. The lower number of stabilizing interactions between the pentamers and the lack of formation of new holes support this model. This model differs from the currently accepted model for rhinoviruses and enteroviruses, in which genome externalization occurs by extrusion of the RNA through capsid channels.
Organizational Affiliation:
Instituto Biofisika (CSIC, UPV/EHU), Barrio Sarriena S/N, 48940 Leioa, Bizkaia, Spain.