Crystal structure of MGS-MChE2, an alpha/beta hydrolase enzyme from the metagenome of sediments from the lagoon of Mar Chica, Morocco
Martinez-Martinez, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MGS-MChE2 | 254 | uncultured bacterium | Mutation(s): 0 | 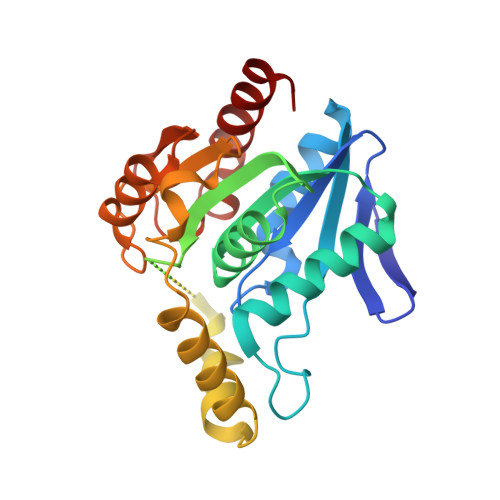 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.968 | α = 90 |
| b = 78.968 | β = 90 |
| c = 78.384 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHENIX | phasing |
| PHENIX | model building |
| Coot | model building |