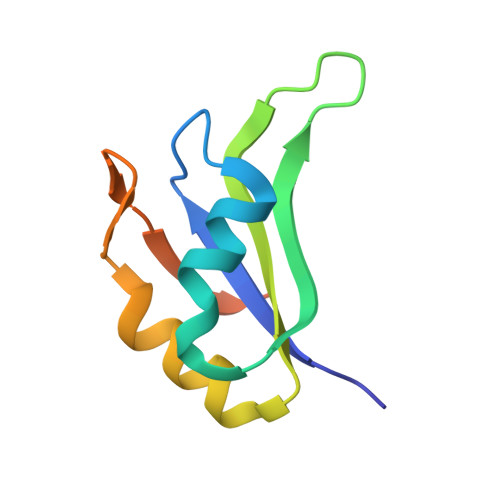
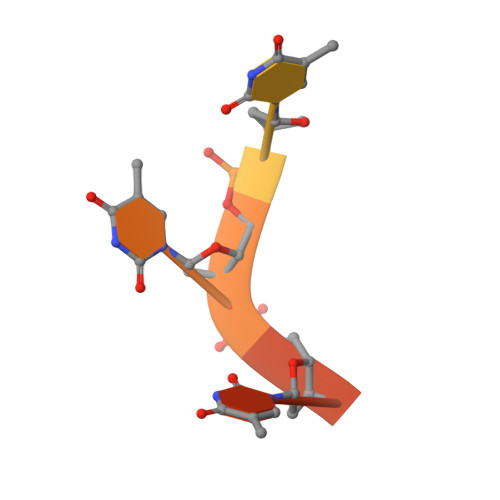
TIA-1 RRM23 binding and recognition of target oligonucleotides.
Waris, S., Garcia-Maurino, S.M., Sivakumaran, A., Beckham, S.A., Loughlin, F.E., Gorospe, M., Diaz-Moreno, I., Wilce, M.C.J., Wilce, J.A.(2017) Nucleic Acids Res 45: 4944-4957
- PubMed: 28184449
- DOI: https://doi.org/10.1093/nar/gkx102
- Primary Citation of Related Structures:
5ITH - PubMed Abstract:
TIA-1 (T-cell restricted intracellular antigen-1) is an RNA-binding protein involved in splicing and translational repression. It mainly interacts with RNA via its second and third RNA recognition motifs (RRMs), with specificity for U-rich sequences directed by RRM2. It has recently been shown that RRM3 also contributes to binding, with preferential binding for C-rich sequences. Here we designed UC-rich and CU-rich 10-nt sequences for engagement of both RRM2 and RRM3 and demonstrated that the TIA-1 RRM23 construct preferentially binds the UC-rich RNA ligand (5΄-UUUUUACUCC-3΄). Interestingly, this binding depends on the presence of Lys274 that is C-terminal to RRM3 and binding to equivalent DNA sequences occurs with similar affinity. Small-angle X-ray scattering was used to demonstrate that, upon complex formation with target RNA or DNA, TIA-1 RRM23 adopts a compact structure, showing that both RRMs engage with the target 10-nt sequences to form the complex. We also report the crystal structure of TIA-1 RRM2 in complex with DNA to 2.3 Å resolution providing the first atomic resolution structure of any TIA protein RRM in complex with oligonucleotide. Together our data support a specific mode of TIA-1 RRM23 interaction with target oligonucleotides consistent with the role of TIA-1 in binding RNA to regulate gene expression.
Organizational Affiliation:
Monash Biomedicine Discovery Institute, Department of Biochemistry & Molecular Biology, Monash University, Victoria 3800, Australia.