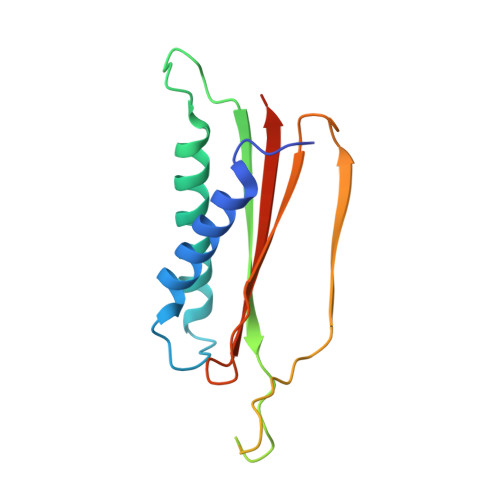
Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Li, G., Huang, J., Yang, J., He, D., Wang, C., Qi, X., Taylor, I.A., Liu, J., Peng, Y.L.(2018) Sci Rep 8: 3952-3952
- PubMed: 29500373
- DOI: https://doi.org/10.1038/s41598-018-21667-5
- Primary Citation of Related Structures:
5HXL, 5HYC - PubMed Abstract:
Magnaporthe oryzae is a model fungal plant pathogen employed for studying plant-fungi interactions. Whole genome sequencing and bioinformatics analyses revealed that this fungal pathogen has more than 12,000 protein-coding genes with 65% of the genes remaining functionally un-annotated. Here, we determine the structure of the hypothetical protein, MGG_01005 and show that it is the Magnaporthe oryzae Dynein light chain Tctex-type 1 (dynlt1/3), demonstrated by its structural similarity to other orthologous dynlt1 proteins and its conserved interaction with the N-terminus of the Magnaporthe oryzae dynein intermediate chain, MoDyn1I2. In addition, we present the structure of the MGG_01005-MoDyn1I2 complex together with mutagenesis studies that reveals a di-histidine motif interaction with a glutamate residue in the dynein intermediate chain within a conserved molecular interface. These results demonstrate the utility of structure-based annotation and validate it as a viable approach for the molecular assignment of hypothetic proteins from phyto-pathogenic fungi.
Organizational Affiliation:
MOA Key Laboratory of Plant Pathology, China Agricultural University, No2 Yunamingyuanxilu, Beijing, 100193, China.