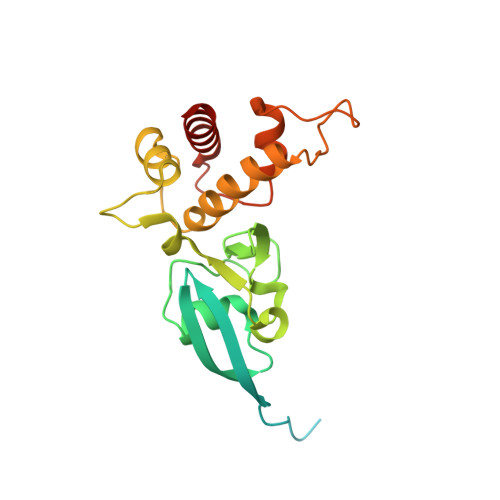
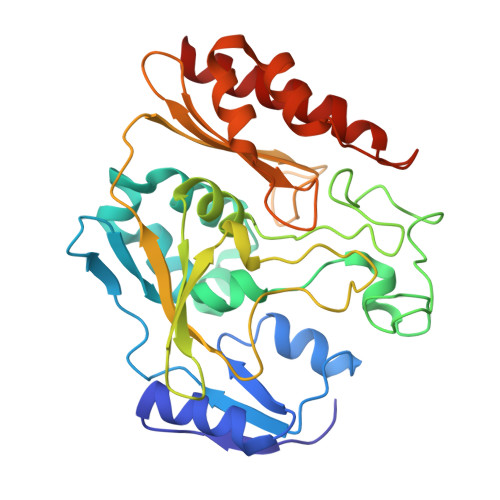
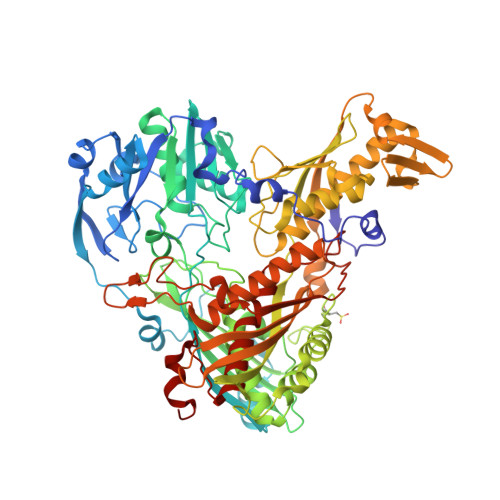
The Escherichia Coli Periplasmic Aldehyde Oxidoreductase is an Exceptional Member of the Xanthine Oxidase Family of Molybdoenzymes.
Correia, M.A., Otrelo-Cardoso, A.R., Schwuchow, V., Sigfridsson Clauss, K.G., Haumann, M., Romao, M.J., Leimkuhler, S., Santos-Silva, T.(2016) ACS Chem Biol 11: 2923
- PubMed: 27622978
- DOI: https://doi.org/10.1021/acschembio.6b00572
- Primary Citation of Related Structures:
5G5G, 5G5H - PubMed Abstract:
The xanthine oxidase (XO) family comprises molybdenum-dependent enzymes that usually form homodimers (or dimers of heterodimers/trimers) organized in three domains that harbor two [2Fe-2S] clusters, one FAD, and a Mo cofactor. In this work, we crystallized an unusual member of the family, the periplasmic aldehyde oxidoreductase PaoABC from Escherichia coli. This is the first example of an E. coli protein containing a molybdopterin-cytosine-dinucleotide cofactor and is the only heterotrimer of the XO family so far structurally characterized. The crystal structure revealed the presence of an unexpected [4Fe-4S] cluster, anchored to an additional 40 residues subdomain. According to phylogenetic analysis, proteins containing this cluster are widely spread in many bacteria phyla, putatively through repeated gene transfer events. The active site of PaoABC is highly exposed to the surface with no aromatic residues and an arginine (PaoC-R440) making a direct interaction with PaoC-E692, which acts as a base catalyst. In order to understand the importance of R440, kinetic assays were carried out, and the crystal structure of the PaoC-R440H variant was also determined.
Organizational Affiliation:
UCIBIO/Requimte, Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa , 2829-516 Caparica, Portugal.