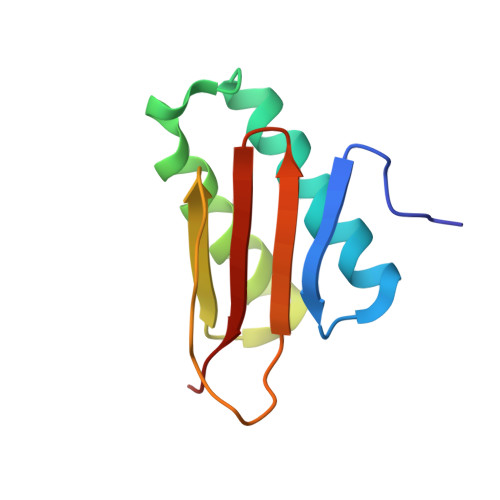
Fasciola hepatica calcium-binding protein FhCaBP2: structure of the dynein light chain-like domain.
Nguyen, T.H., Thomas, C.M., Timson, D.J., van Raaij, M.J.(2016) Parasitol Res 115: 2879-2886
- PubMed: 27083189
- DOI: https://doi.org/10.1007/s00436-016-5046-x
- Primary Citation of Related Structures:
5FWZ, 5FX0 - PubMed Abstract:
The common liver fluke Fasciola hepatica causes an increasing burden on human and animal health, partly because of the spread of drug-resistant isolates. As a consequence, there is considerable interest in developing new drugs to combat liver fluke infections. A group of potential targets is a family of calcium-binding proteins which combine an N-terminal domain with two EF-hand motifs and a C-terminal domain with predicted similarity to dynein light chains (DLC-like domain). The function of these proteins is unknown, although in several species, they have been localised to the tegument, an important structure at the host-parasite interface. Here, we report the X-ray crystal structure of the DLC-like domain of F. hepatica calcium-binding protein 2 (FhCaBP2), solved using single-wavelength anomalous diffraction and refined at 2.3 Å resolution in two different crystal forms. The FhCaBP2 DLC-like domain has a structure similar to other DLC domains, with an anti-parallel β-sheet packed against an α-helical hairpin. Like other DLC domains, it dimerises through its β2-strand, which extends in an arch and forms the fifth strand in an extended β-sheet of the other monomer. The structure provides molecular details of the dimerisation of FhCaBP2, the first example from this family of parasite proteins.
Organizational Affiliation:
Dpto de Estructura de Macromoleculas, Centro Nacional de Biotecnologia-CSIC, calle Darwin 3, E-28049, Madrid, Spain.