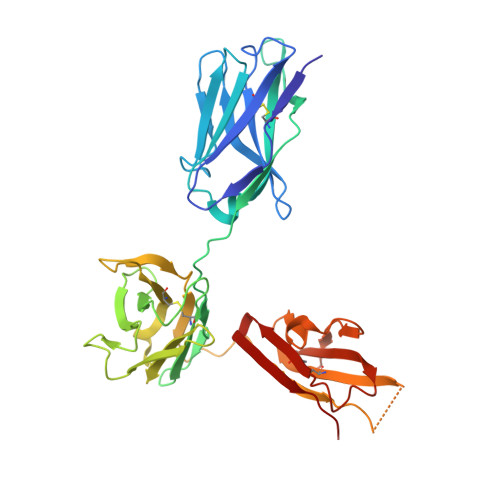
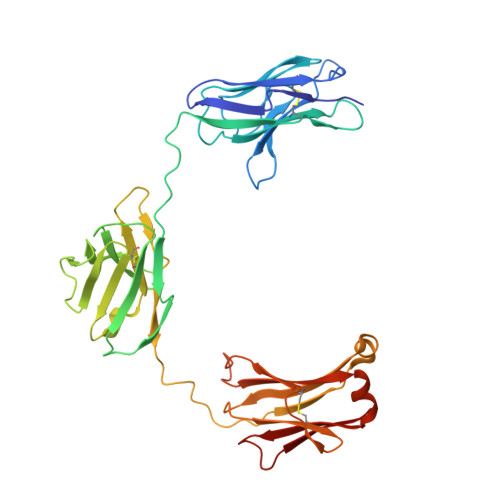
CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Steinmetz, A., Vallee, F., Beil, C., Lange, C., Baurin, N., Beninga, J., Capdevila, C., Corvey, C., Dupuy, A., Ferrari, P., Rak, A., Wonerow, P., Kruip, J., Mikol, V., Rao, E.(2016) MAbs 8: 867-878
- PubMed: 26984268
- DOI: https://doi.org/10.1080/19420862.2016.1162932
- Primary Citation of Related Structures:
5FHX, 5HCG - PubMed Abstract:
Bispecific immunoglobulins (Igs) typically contain at least two distinct variable domains (Fv) that bind to two different target proteins. They are conceived to facilitate clinical development of biotherapeutic agents for diseases where improved clinical outcome is obtained or expected by combination therapy compared to treatment by single agents. Almost all existing formats are linear in their concept and differ widely in drug-like and manufacture-related properties. To overcome their major limitations, we designed cross-over dual variable Ig-like proteins (CODV-Ig). Their design is akin to the design of circularly closed repeat architectures. Indeed, initial results showed that the traditional approach of utilizing (G4S)x linkers for biotherapeutics design does not identify functional CODV-Igs. Therefore, we applied an unprecedented molecular modeling strategy for linker design that consistently results in CODV-Igs with excellent biochemical and biophysical properties. CODV architecture results in a circular self-contained structure functioning as a self-supporting truss that maintains the parental antibody affinities for both antigens without positional effects. The format is universally suitable for therapeutic applications targeting both circulating and membrane-localized proteins. Due to the full functionality of the Fc domains, serum half-life extension as well as antibody- or complement-dependent cytotoxicity may support biological efficiency of CODV-Igs. We show that judicious choice in combination of epitopes and paratope orientations of bispecific biotherapeutics is anticipated to be critical for clinical outcome. Uniting the major advantages of alternative bispecific biotherapeutics, CODV-Igs are applicable in a wide range of disease areas for fast-track multi-parametric drug optimization.
Organizational Affiliation:
c Sanofi R&D, LGCR, Center de Recherche Vitry-sur-Seine , Vitry-sur-Seine Cedex , France.