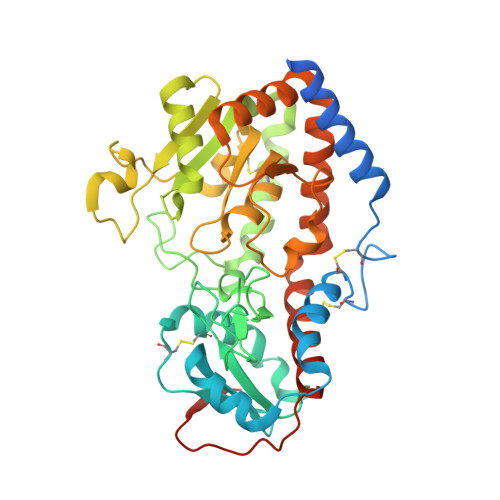
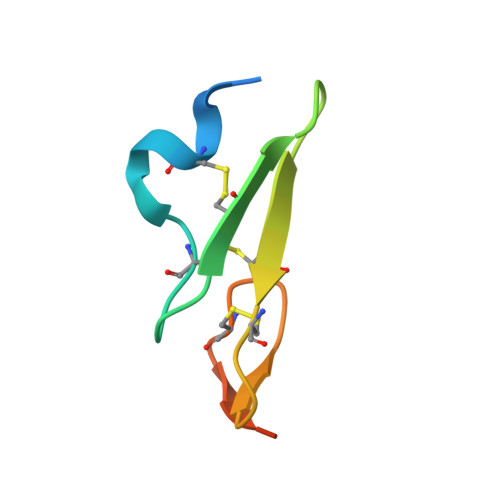
Structural analysis of Notch-regulating Rumi reveals basis for pathogenic mutations.
Yu, H., Takeuchi, H., Takeuchi, M., Liu, Q., Kantharia, J., Haltiwanger, R.S., Li, H.(2016) Nat Chem Biol 12: 735-740
- PubMed: 27428513
- DOI: https://doi.org/10.1038/nchembio.2135
- Primary Citation of Related Structures:
5F84, 5F85, 5F86, 5F87 - PubMed Abstract:
Rumi O-glucosylates the EGF repeats of a growing list of proteins essential in metazoan development, including Notch. Rumi is essential for Notch signaling, and Rumi dysregulation is linked to several human diseases. Despite Rumi's critical roles, it is unknown how Rumi glucosylates a serine of many but not all EGF repeats. Here we report crystal structures of Drosophila Rumi as binary and ternary complexes with a folded EGF repeat and/or donor substrates. These structures provide insights into the catalytic mechanism and show that Rumi recognizes structural signatures of the EGF motif, the U-shaped consensus sequence, C-X-S-X-(P/A)-C and a conserved hydrophobic region. We found that five Rumi mutations identified in cancers and Dowling-Degos disease are clustered around the enzyme active site and adversely affect its activity. Our study suggests that loss of Rumi activity may underlie these diseases, and the mechanistic insights may facilitate the development of modulators of Notch signaling.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, New York, USA.