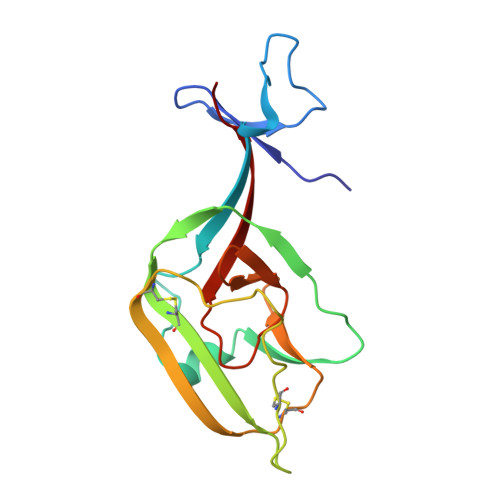
Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Wang, H., Shi, Y., Song, J., Qi, J., Lu, G., Yan, J., Gao, G.F.(2016) Cell 164: 258-268
- PubMed: 26771495
- DOI: https://doi.org/10.1016/j.cell.2015.12.044
- Primary Citation of Related Structures:
5F18, 5F1B - PubMed Abstract:
Filoviruses, including Ebola and Marburg, cause fatal hemorrhagic fever in humans and primates. Understanding how these viruses enter host cells could help to develop effective therapeutics. An endosomal protein, Niemann-Pick C1 (NPC1), has been identified as a necessary entry receptor for this process, and priming of the viral glycoprotein (GP) to a fusion-competent state is a prerequisite for NPC1 binding. Here, we have determined the crystal structure of the primed GP (GPcl) of Ebola virus bound to domain C of NPC1 (NPC1-C) at a resolution of 2.3 Å. NPC1-C utilizes two protruding loops to engage a hydrophobic cavity on head of GPcl. Upon enzymatic cleavage and NPC1-C binding, conformational change in the GPcl further affects the state of the internal fusion loop, triggering membrane fusion. Our data therefore provide structural insights into filovirus entry in the late endosome and the molecular basis for design of therapeutic inhibitors of viral entry.
Organizational Affiliation:
CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.