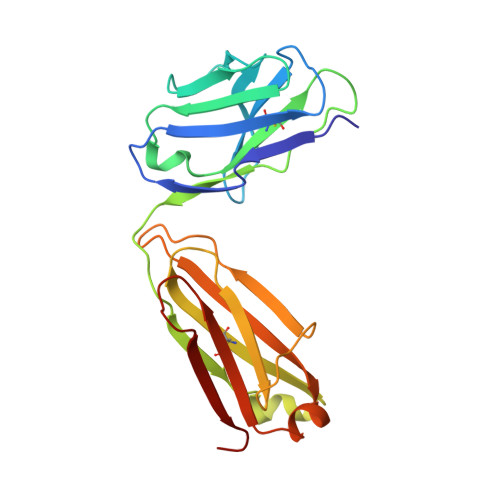
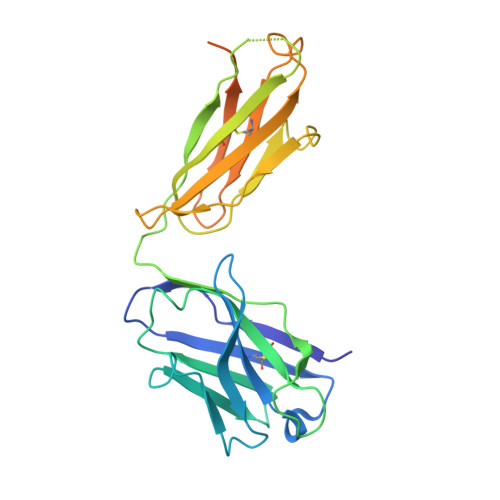
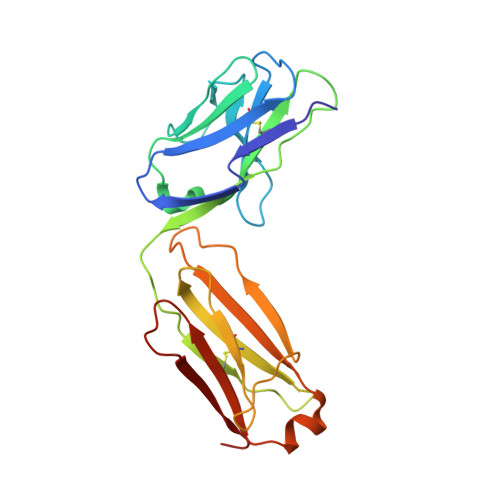
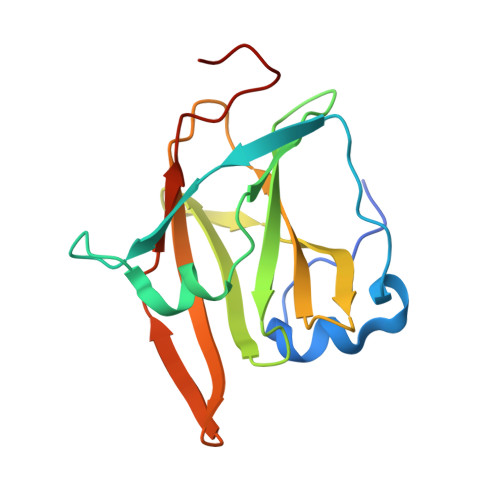
Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire.
Yeung, Y.A., Foletti, D., Deng, X., Abdiche, Y., Strop, P., Glanville, J., Pitts, S., Lindquist, K., Sundar, P.D., Sirota, M., Hasa-Moreno, A., Pham, A., Melton Witt, J., Ni, I., Pons, J., Shelton, D., Rajpal, A., Chaparro-Riggers, J.(2016) Nat Commun 7: 13376-13376
- PubMed: 27857134
- DOI: https://doi.org/10.1038/ncomms13376
- Primary Citation of Related Structures:
5D1Q, 5D1X, 5D1Z - PubMed Abstract:
Staphylococcus aureus is both an important pathogen and a human commensal. To explore this ambivalent relationship between host and microbe, we analysed the memory humoral response against IsdB, a protein involved in iron acquisition, in four healthy donors. Here we show that in all donors a heavily biased use of two immunoglobulin heavy chain germlines generated high affinity (pM) antibodies that neutralize the two IsdB NEAT domains, IGHV4-39 for NEAT1 and IGHV1-69 for NEAT2. In contrast to the typical antibody/antigen interactions, the binding is primarily driven by the germline-encoded hydrophobic CDRH-2 motifs of IGHV1-69 and IGHV4-39, with a binding mechanism nearly identical for each antibody derived from different donors. Our results suggest that IGHV1-69 and IGHV4-39, while part of the adaptive immune system, may have evolved under selection pressure to encode a binding motif innately capable of recognizing and neutralizing a structurally conserved protein domain involved in pathogen iron acquisition.
Organizational Affiliation:
Rinat R&D, Pfizer Inc., 230 East Grand Avenue, South San Francisco, California 94080, USA.