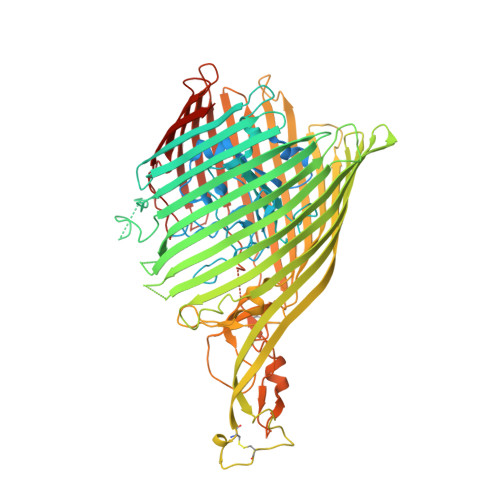
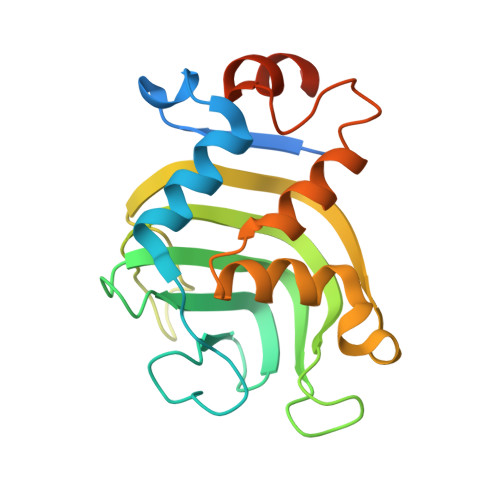
Binding of HasA by its transmembrane receptor HasR follows a conformational funnel mechanism.
Exner, T.E., Becker, S., Becker, S., Boniface-Guiraud, A., Delepelaire, P., Diederichs, K., Welte, W.(2020) Eur Biophys J 49: 39-57
- PubMed: 31802151
- DOI: https://doi.org/10.1007/s00249-019-01411-1
- Primary Citation of Related Structures:
5C58 - PubMed Abstract:
HasR in the outer membrane of Serratia marcescens binds secreted, heme-loaded HasA and translocates the heme to the periplasm to satisfy the cell's demand for iron. The previously published crystal structure of the wild-type complex showed HasA in a very specific binding arrangement with HasR, apt to relax the grasp on the heme and assure its directed transfer to the HasR-binding site. Here, we present a new crystal structure of the heme-loaded HasA arranged with a mutant of HasR, called double mutant (DM) in the following that seemed to mimic a precursor stage of the abovementioned final arrangement before heme transfer. To test this, we performed first molecular dynamics (MD) simulations starting at the crystal structure of the complex of HasA with the DM mutant and then targeted MD simulations of the entire binding process beginning with heme-loaded HasA in solution. When the simulation starts with the former complex, the two proteins in most simulations do not dissociate. When the mutations are reverted to the wild-type sequence, dissociation and development toward the wild-type complex occur in most simulations. This indicates that the mutations create or enhance a local energy minimum. In the targeted MD simulations, the first protein contacts depend upon the chosen starting position of HasA in solution. Subsequently, heme-loaded HasA slides on the external surface of HasR on paths that converge toward the specific arrangement apt for heme transfer. The targeted simulations end when HasR starts to relax the grasp on the heme, the subsequent events being in a time regime inaccessible to the available computing power. Interestingly, none of the ten independent simulation paths visits exactly the arrangement of HasA with HasR seen in the crystal structure of the mutant. Two factors which do not exclude each other could explain these observations: the double mutation creates a non-physiologic potential energy minimum between the two proteins and /or the target potential in the simulation pushes the system along paths deviating from the low-energy paths of the native binding processes. Our results support the former view, but do not exclude the latter possibility.
Organizational Affiliation:
Pharmazeutisches Institut, University of Tübingen, Auf der Morgenstelle 8, 72076, Tübingen, Germany.