A Guide to Fluorescent Protein FRET Pairs.
Bajar, B.T., Wang, E.S., Zhang, S., Lin, M.Z., Chu, J.(2016) Sensors (Basel) 16
- PubMed: 27649177
- DOI: https://doi.org/10.3390/s16091488
- Primary Citation of Related Structures:
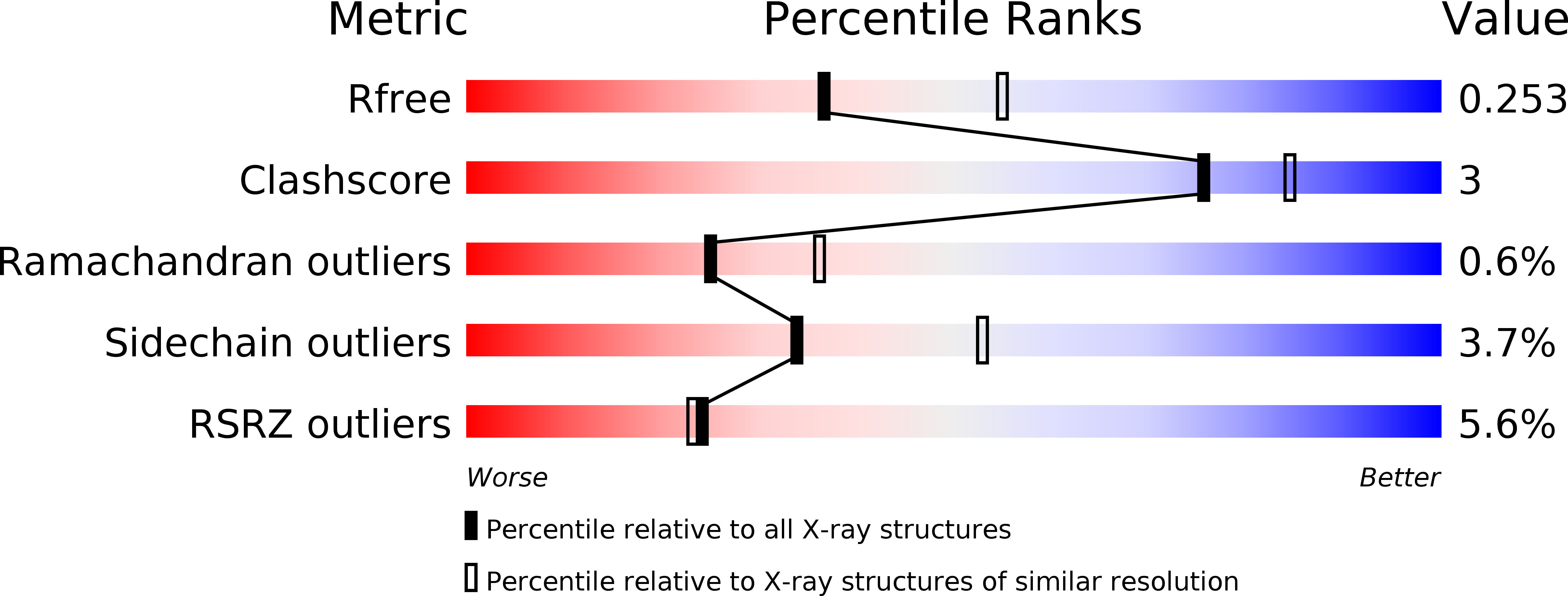
5BQL - PubMed Abstract:
Förster or fluorescence resonance energy transfer (FRET) technology and genetically encoded FRET biosensors provide a powerful tool for visualizing signaling molecules in live cells with high spatiotemporal resolution. Fluorescent proteins (FPs) are most commonly used as both donor and acceptor fluorophores in FRET biosensors, especially since FPs are genetically encodable and live-cell compatible. In this review, we will provide an overview of methods to measure FRET changes in biological contexts, discuss the palette of FP FRET pairs developed and their relative strengths and weaknesses, and note important factors to consider when using FPs for FRET studies.
Organizational Affiliation:
Medical Scientist Training Program, University of California, Los Angeles, CA 90095, USA. BBajar@mednet.ucla.edu.