Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense
Fischer, A., Harrison, K.S., Ramirez, Y., Auer, D., Chowdhury, S.R., Prusty, B.K., Sauer, F., Dimond, Z., Kisker, C., Scott Hefty, P., Rudel, T.(2017) Elife 6
- PubMed: 28347402
- DOI: https://doi.org/10.7554/eLife.21465
- Primary Citation of Related Structures:
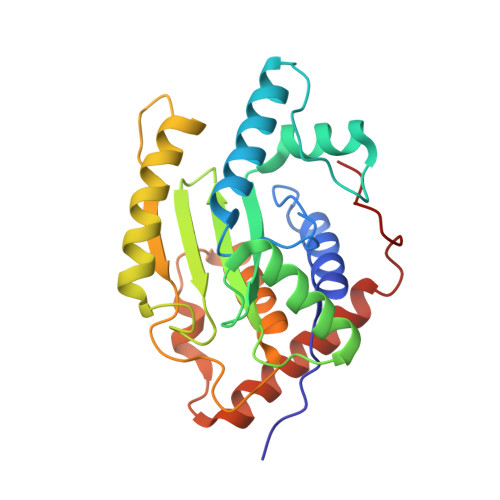
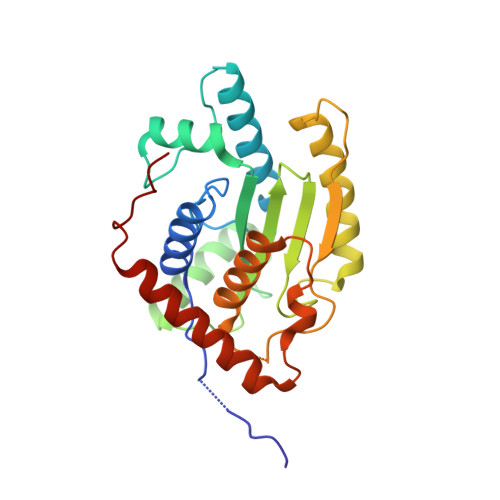
5B5Q - PubMed Abstract:
Obligate intracellular Chlamydia trachomatis replicate in a membrane-bound vacuole called inclusion, which serves as a signaling interface with the host cell. Here, we show that the chlamydial deubiquitinating enzyme (Cdu) 1 localizes in the inclusion membrane and faces the cytosol with the active deubiquitinating enzyme domain. The structure of this domain revealed high similarity to mammalian deubiquitinases with a unique α-helix close to the substrate-binding pocket. We identified the apoptosis regulator Mcl-1 as a target that interacts with Cdu1 and is stabilized by deubiquitination at the chlamydial inclusion. A chlamydial transposon insertion mutant in the Cdu1-encoding gene exhibited increased Mcl-1 and inclusion ubiquitination and reduced Mcl-1 stabilization. Additionally, inactivation of Cdu1 led to increased sensitivity of C. trachomatis for IFNγ and impaired infection in mice. Thus, the chlamydial inclusion serves as an enriched site for a deubiquitinating activity exerting a function in selective stabilization of host proteins and protection from host defense.
Organizational Affiliation:
Department of Microbiology, Biocenter, University of Würzburg, Würzburg, Germany.