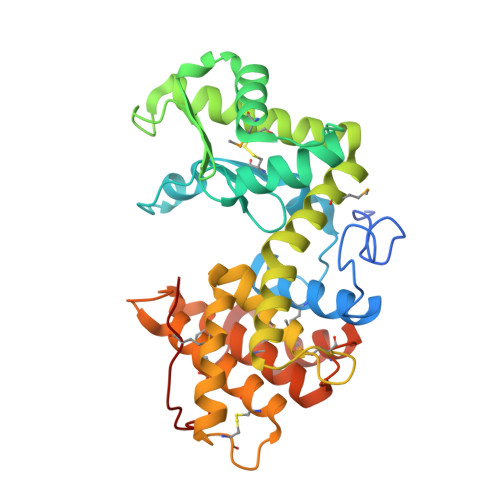
Crystal structure of a family 80 chitosanase from Mitsuaria chitosanitabida
Yorinaga, Y., Kumasaka, T., Yamamoto, M., Hamada, K., Kawamukai, M.(2017) FEBS Lett 591: 540-547
- PubMed: 28084023
- DOI: https://doi.org/10.1002/1873-3468.12557
- Primary Citation of Related Structures:
5B4S - PubMed Abstract:
Chitosanases belong to glycoside hydrolase families 5, 7, 8, 46, 75 and 80 and hydrolyse glucosamine polymers produced by partial or full deacetylation of chitin. Herein, we determined the crystal structure of chitosanase from the β-proteobacterium, Mitsuaria chitosanitabida, (McChoA) at 1.75 Å resolution; the first structure of a family 80 chitosanase. McChoA is a 34 kDa extracellular protein of 301 amino acids that fold into two (upper and lower) globular domains with an active site cleft between them. Key substrate-binding features are conserved with family 24 lysozymes and family 46 chitosanases. The distance between catalytic residues E41 and E61 (10.8 Å) indicates an inverting type mechanism. Uniquely, three disulphide bridges and the C terminus might contribute to enzyme activity.
Organizational Affiliation:
Department of Life Science and Biotechnology, Faculty of Life and Environmental Science, Shimane University, Matsue, Japan.