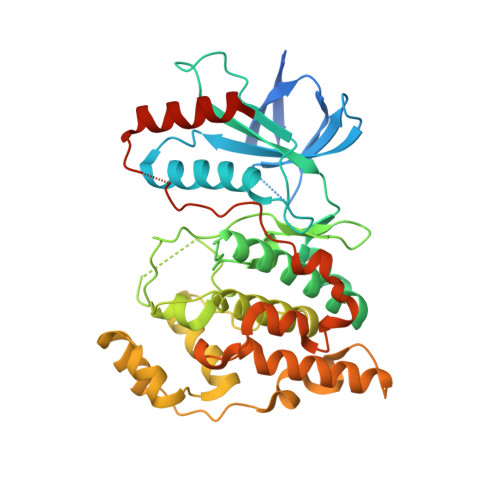
The crystal structure of JNK from Drosophila melanogaster reveals an evolutionarily conserved topology with that of mammalian JNK proteins.
Chimnaronk, S., Sitthiroongruang, J., Srisucharitpanit, K., Srisaisup, M., Ketterman, A.J., Boonserm, P.(2015) BMC Struct Biol 15: 17-17
- PubMed: 26377800
- DOI: https://doi.org/10.1186/s12900-015-0045-1
- Primary Citation of Related Structures:
5AWM - PubMed Abstract:
The c-Jun N-terminal kinases (JNKs), members of the mitogen-activated protein kinase (MAPK) family, engage in diverse cellular responses to signals produced under normal development and stress conditions. In Drosophila, only one JNK member is present, whereas ten isoforms from three JNK genes (JNK1, 2, and 3) are present in mammalian cells. To date, several mammalian JNK structures have been determined, however, there has been no report of any insect JNK structure.
Organizational Affiliation:
Institute of Molecular Biosciences, Mahidol University, Salaya, Phuttamonthon, Nakhon Pathom, 73170, Thailand. sarin.chi@mahidol.ac.th.