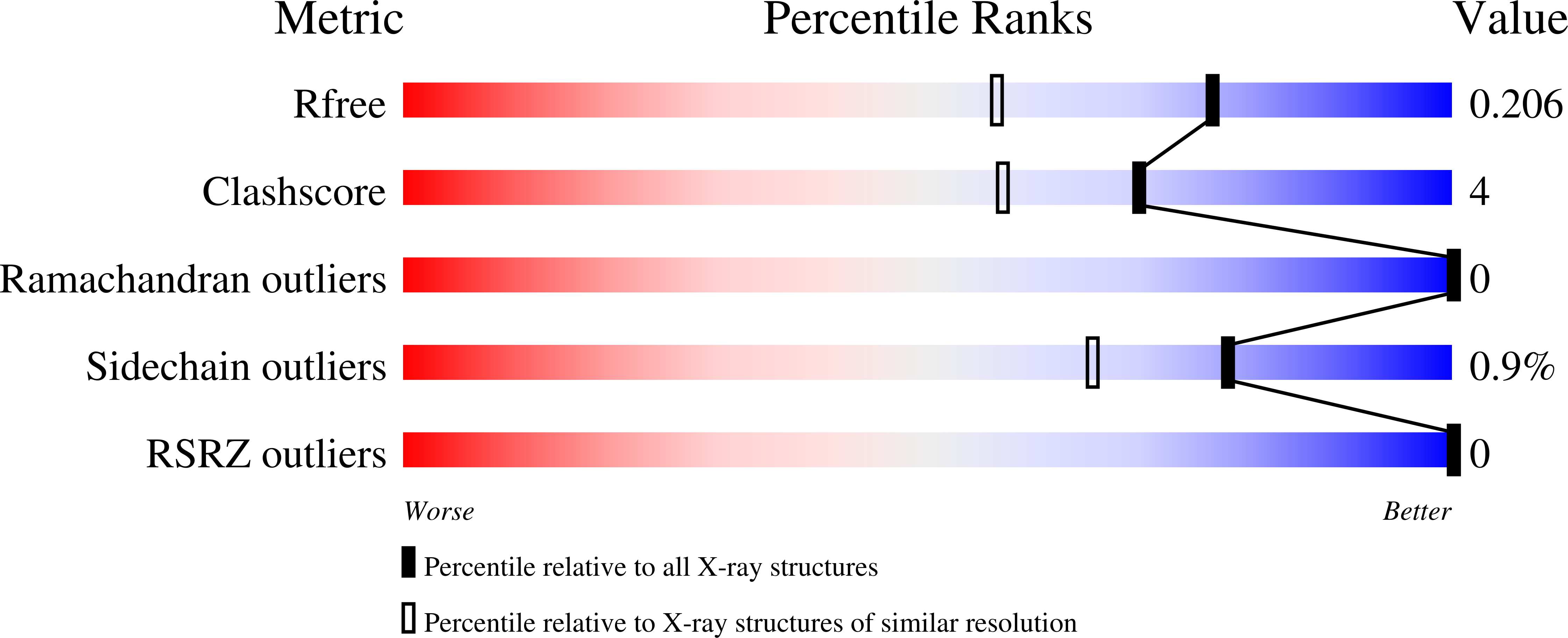
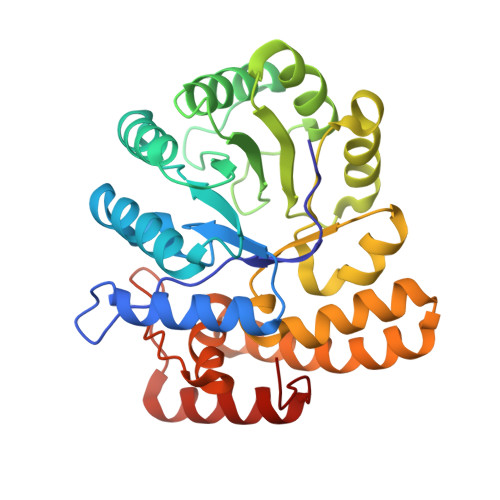
Features and structure of a cold active N-acetylneuraminate lyase.
Gurung, M.K., Altermark, B., Helland, R., Smalas, A.O., Raeder, I.L.U.(2019) PLoS One 14: e0217713-e0217713
- PubMed: 31185017
- DOI: https://doi.org/10.1371/journal.pone.0217713
- Primary Citation of Related Structures:
5AFD - PubMed Abstract:
N-acetylneuraminate lyases (NALs) are enzymes that catalyze the reversible cleavage and synthesis of sialic acids. They are therefore commonly used for the production of these high-value sugars. This study presents the recombinant production, together with biochemical and structural data, of the NAL from the psychrophilic bacterium Aliivibrio salmonicida LFI1238 (AsNAL). Our characterization shows that AsNAL possesses high activity and stability at alkaline pH. We confirm that these properties allow for the use in a one-pot reaction at alkaline pH for the synthesis of N-acetylneuraminic acid (Neu5Ac, the most common sialic acid) from the inexpensive precursor N-acetylglucosamine. We also show that the enzyme has a cold active nature with an optimum temperature for Neu5Ac synthesis at 20°C. The equilibrium constant for the reaction was calculated at different temperatures, and the formation of Neu5Ac acid is favored at low temperatures, making the cold active enzyme a well-suited candidate for use in such exothermic reactions. The specific activity is high compared to the homologue from Escherichia coli at three tested temperatures, and the enzyme shows a higher catalytic efficiency and turnover number for cleavage at 37°C. Mutational studies reveal that amino acid residue Asn 168 is important for the high kcat. The crystal structure of AsNAL was solved to 1.65 Å resolution and reveals a compact, tetrameric protein similar to other NAL structures. The data presented provides a framework to guide further optimization of its application in sialic acid production and opens the possibility for further design of the enzyme.
Organizational Affiliation:
The Norwegian Structural Biology Center (NorStruct), Department of Chemistry, UiT- The Arctic University of Norway, Tromsø, Norway.