Deep Sequencing-guided Design of a High Affinity Dual Specificity Antibody to Target Two Angiogenic Factors in Neovascular Age-related Macular Degeneration.
Koenig, P., Lee, C.V., Sanowar, S., Wu, P., Stinson, J., Harris, S.F., Fuh, G.(2015) J Biol Chem 290: 21773-21786
- PubMed: 26088137
- DOI: https://doi.org/10.1074/jbc.M115.662783
- Primary Citation of Related Structures:
4ZFF, 4ZFG - PubMed Abstract:
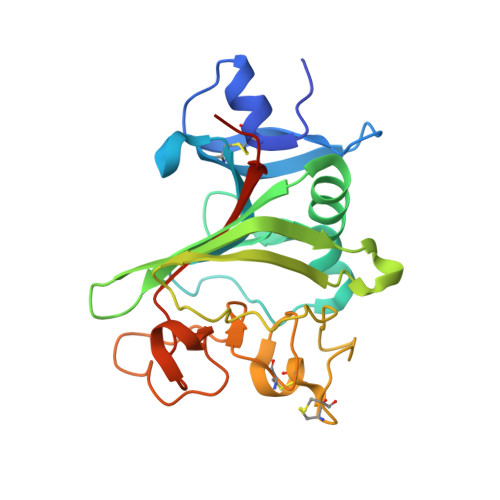
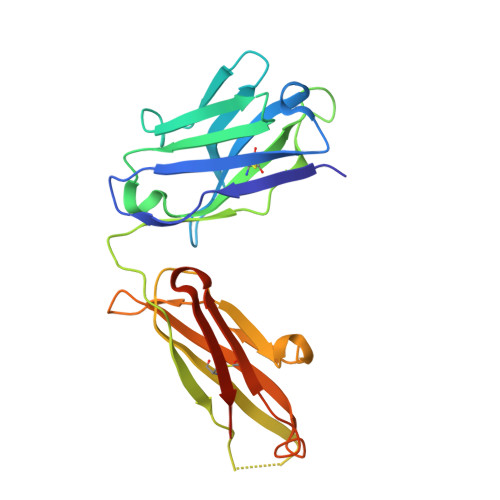
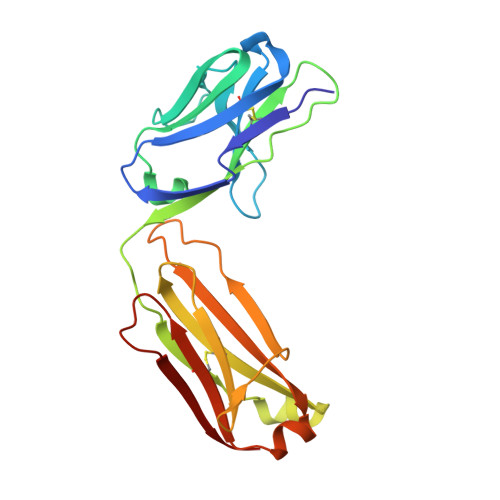
The development of dual targeting antibodies promises therapies with improved efficacy over mono-specific antibodies. Here, we engineered a Two-in-One VEGF/angiopoietin 2 antibody with dual action Fab (DAF) as a potential therapeutic for neovascular age-related macular degeneration. Crystal structures of the VEGF/angiopoietin 2 DAF in complex with its two antigens showed highly overlapping binding sites. To achieve sufficient affinity of the DAF to block both angiogenic factors, we turned to deep mutational scanning in the complementarity determining regions (CDRs). By mutating all three CDRs of each antibody chain simultaneously, we were able not only to identify affinity improving single mutations but also mutation pairs from different CDRs that synergistically improve both binding functions. Furthermore, insights into the cooperativity between mutations allowed us to identify fold-stabilizing mutations in the CDRs. The data obtained from deep mutational scanning reveal that the majority of the 52 CDR residues are utilized differently for the two antigen binding function and permit, for the first time, the engineering of several DAF variants with sub-nanomolar affinity against two structurally unrelated antigens. The improved variants show similar blocking activity of receptor binding as the high affinity mono-specific antibodies against these two proteins, demonstrating the feasibility of generating a dual specificity binding surface with comparable properties to individual high affinity mono-specific antibodies.
Organizational Affiliation:
From the Departments of Antibody Engineering.