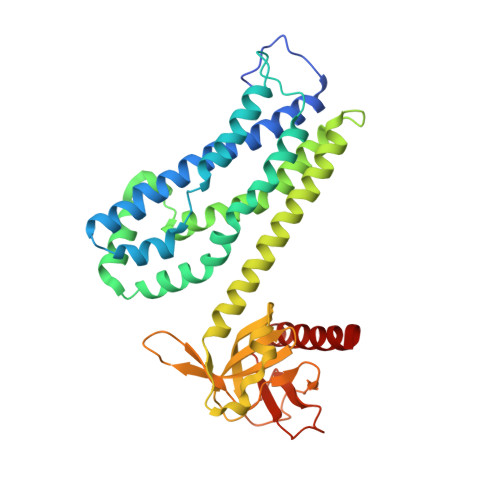
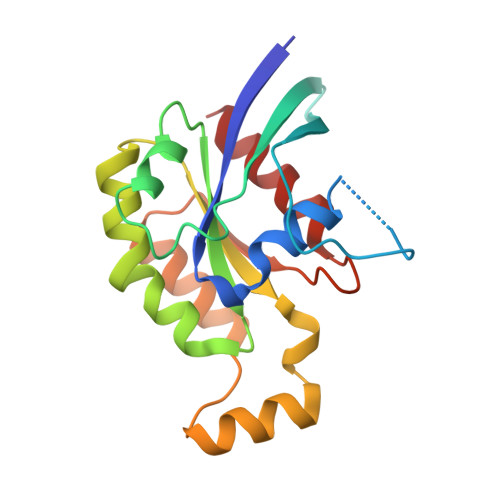
A structural study of the complex between neuroepithelial cell transforming gene 1 (Net1) and RhoA reveals a potential anticancer drug hot spot.
Petit, A.P., Garcia-Petit, C., Bueren-Calabuig, J.A., Vuillard, L.M., Ferry, G., Boutin, J.A.(2018) J Biol Chem 293: 9064-9077
- PubMed: 29695506
- DOI: https://doi.org/10.1074/jbc.RA117.001123
- Primary Citation of Related Structures:
4XH9 - PubMed Abstract:
The GTPase RhoA is a major player in many different regulatory pathways. RhoA catalyzes GTP hydrolysis, and its catalysis is accelerated when RhoA forms heterodimers with proteins of the guanine nucleotide exchange factor (GEF) family. Neuroepithelial cell transforming gene 1 (Net1) is a RhoA-interacting GEF implicated in cancer, but the structural features supporting the RhoA/Net1 interaction are unknown. Taking advantage of a simple production and purification process, here we solved the structure of a RhoA/Net1 heterodimer with X-ray crystallography at 2-Å resolution. Using a panel of several techniques, including molecular dynamics simulations, we characterized the RhoA/Net1 interface. Moreover, deploying an extremely simple peptide-based scanning approach, we found that short peptides (penta- to nonapeptides) derived from the protein/protein interaction region of RhoA could disrupt the RhoA/Net1 interaction and thereby diminish the rate of nucleotide exchange. The most inhibitory peptide, EVKHF, spanning residues 102-106 in the RhoA sequence, displayed an IC 50 of ∼100 μm without further modifications. The peptides identified here could be useful in further investigations of the RhoA/Net1 interaction region. We propose that our structural and functional insights might inform chemical approaches for transforming the pentapeptide into an optimized pseudopeptide that antagonizes Net1-mediated RhoA activation with therapeutic anticancer potential.
Organizational Affiliation:
From the Drug Discovery Unit, Division of Biological Chemistry and Drug Discovery and.