Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Liu, J., Huang, J., Zhao, Y., Liu, H., Wang, D., Yang, J., Zhao, W., Taylor, I.A., Peng, Y.(2015) Nucleic Acids Res 43: 1231
- PubMed: 25550425
- DOI: https://doi.org/10.1093/nar/gku1351
- Primary Citation of Related Structures:
4UX5 - PubMed Abstract:
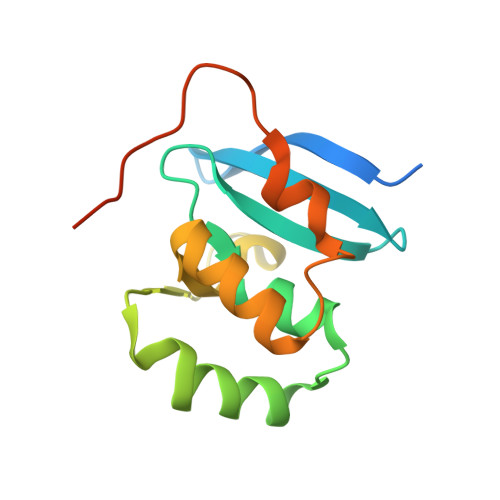
The MBP1 family proteins are the DNA binding subunits of MBF cell-cycle transcription factor complexes and contain an N terminal winged helix-turn-helix (wHTH) DNA binding domain (DBD). Although the DNA binding mechanism of MBP1 from Saccharomyces cerevisiae has been extensively studied, the structural framework and the DNA binding mode of other MBP1 family proteins remains to be disclosed. Here, we determined the crystal structure of the DBD of PCG2, the Magnaporthe oryzae orthologue of MBP1, bound to MCB-DNA. The structure revealed that the wing, the 20-loop, helix A and helix B in PCG2-DBD are important elements for DNA binding. Unlike previously characterized wHTH proteins, PCG2-DBD utilizes the wing and helix-B to bind the minor groove and the major groove of the MCB-DNA whilst the 20-loop and helix A interact non-specifically with DNA. Notably, two glutamines Q89 and Q82 within the wing were found to recognize the MCB core CGCG sequence through making hydrogen bond interactions. Further in vitro assays confirmed essential roles of Q89 and Q82 in the DNA binding. These data together indicate that the MBP1 homologue PCG2 employs an unusual mode of binding to target DNA and demonstrate the versatility of wHTH domains.
Organizational Affiliation:
MOA Key Laboratory of Plant Pathology, China Agricultural University, Beijing 100193, China.