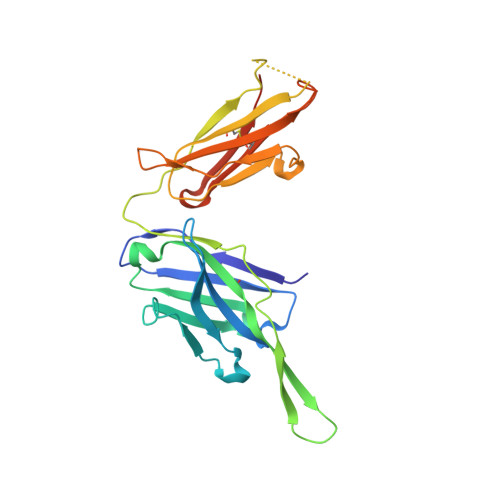
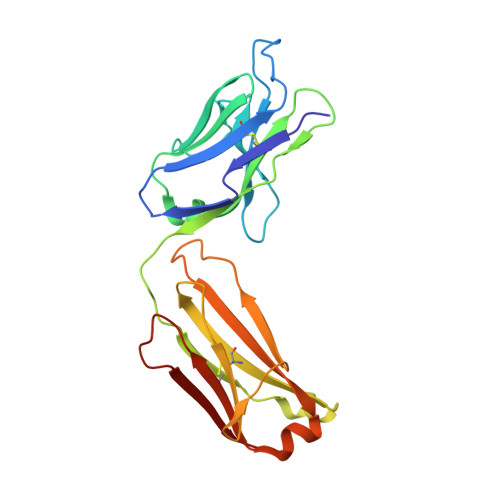
Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Douthwaite, J.A., Sridharan, S., Huntington, C., Hammersley, J., Marwood, R., Hakulinen, J.K., Ek, M., Sjogren, T., Rider, D., Privezentzev, C., Seaman, J.C., Cariuk, P., Knights, V., Young, J., Wilkinson, T., Sleeman, M., Finch, D.K., Lowe, D.C., Vaughan, T.J.(2015) MAbs 7: 152
- PubMed: 25484051
- DOI: https://doi.org/10.4161/19420862.2014.985158
- Primary Citation of Related Structures:
4UV4 - PubMed Abstract:
Therapeutic monoclonal antibodies targeting G-protein-coupled receptors (GPCRs) are desirable for intervention in a wide range of disease processes. The discovery of such antibodies is challenging due to a lack of stability of many GPCRs as purified proteins. We describe here the generation of Fpro0165, a human anti-formyl peptide receptor 1 (FPR1) antibody generated by variable domain engineering of an antibody derived by immunization of transgenic mice expressing human variable region genes. Antibody isolation and subsequent engineering of affinity, potency and species cross-reactivity using phage display were achieved using FPR1 expressed on HEK cells for immunization and selection, along with calcium release cellular assays for antibody screening. Fpro0165 shows full neutralization of formyl peptide-mediated activation of primary human neutrophils. A crystal structure of the Fpro0165 Fab shows a long, protruding VH CDR3 of 24 amino acids and in silico docking with a homology model of FPR1 suggests that this long VH CDR3 is critical to the predicted binding mode of the antibody. Antibody mutation studies identify the apex of the long VH CDR3 as key to mediating the species cross-reactivity profile of the antibody. This study illustrates an approach for antibody discovery and affinity engineering to typically intractable membrane proteins such as GPCRs.
Organizational Affiliation:
a MedImmune Ltd. ; Cambridge , UK.