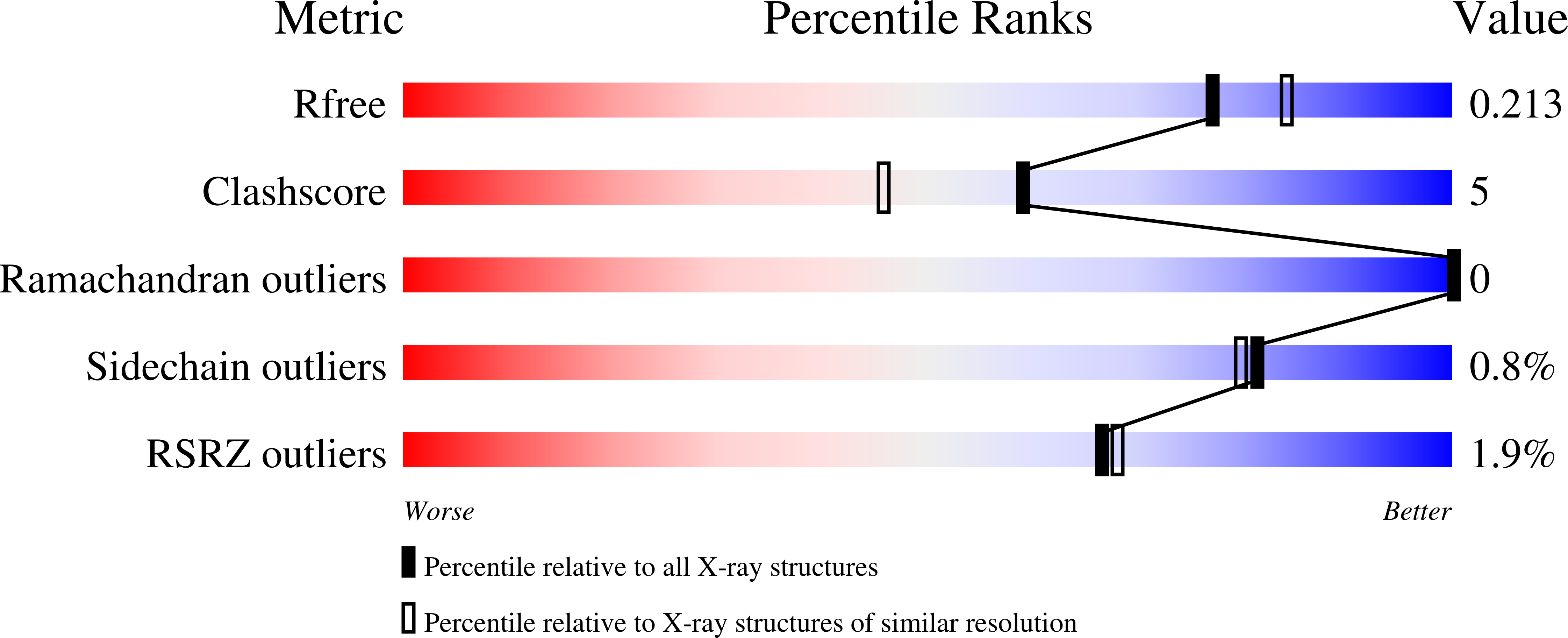
Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
Naik, M., Raichurkar, A., Bandodkar, B.S., Varun, B.V., Bhat, S., Kalkhambkar, R., Murugan, K., Menon, R., Bhat, J., Paul, B., Iyer, H., Hussein, S., Tucker, J.A., Vogtherr, M., Embrey, K.J., Mcmiken, H., Prasad, S., Gill, A., Ugarkar, B.G., Venkatraman, J., Read, J., Panda, M.(2015) J Med Chem 58: 753
- PubMed: 25486447
- DOI: https://doi.org/10.1021/jm5012947
- Primary Citation of Related Structures:
4UNN, 4UNP, 4UNQ, 4UNR, 4UNS - PubMed Abstract:
M. tuberculosis thymidylate kinase (Mtb TMK) has been shown in vitro to be an essential enzyme in DNA synthesis. In order to identify novel leads for Mtb TMK, we performed a high throughput biochemical screen and an NMR based fragment screen through which we discovered two novel classes of inhibitors, 3-cyanopyridones and 1,6-naphthyridin-2-ones, respectively. We describe three cyanopyridone subseries that arose during our hit to lead campaign, along with cocrystal structures of representatives with Mtb TMK. Structure aided optimization of the cyanopyridones led to single digit nanomolar inhibitors of Mtb TMK. Fragment based lead generation, augmented by crystal structures and the SAR from the cyanopyridones, enabled us to drive the potency of our 1,6-naphthyridin-2-one fragment hit from 500 μM to 200 nM while simultaneously improving the ligand efficiency. Cyanopyridone derivatives containing sulfoxides and sulfones showed cellular activity against M. tuberculosis. To the best of our knowledge, these compounds are the first reports of non-thymidine-like inhibitors of Mtb TMK.
Organizational Affiliation:
Department of Chemistry, and ‡Department of Bioscience, Innovative Medicines Infection, AstraZeneca India Pvt. Ltd. , Bellary Road, Hebbal, Bangalore, Karnataka 560024, India.