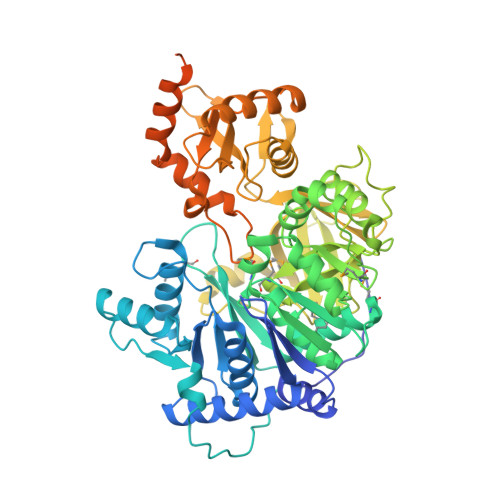
Structure of the adenylation-peptidyl carrier protein didomain of the Microcystis aeruginosa microcystin synthetase McyG.
Tan, X.F., Dai, Y.N., Zhou, K., Jiang, Y.L., Ren, Y.M., Chen, Y., Zhou, C.Z.(2015) Acta Crystallogr D Biol Crystallogr 71: 873-881
- PubMed: 25849398
- DOI: https://doi.org/10.1107/S1399004715001716
- Primary Citation of Related Structures:
4R0M - PubMed Abstract:
Microcystins, which are the most common cause of hepatotoxicity associated with cyanobacterial water blooms, are assembled in vivo on a large multienzyme complex via a mixed nonribosomal peptide synthetase/polyketide synthetase (NRPS/PKS). The biosynthesis of microcystin in Microcystis aeruginosa PCC 7806 starts with the enzyme McyG, which contains an adenylation-peptidyl carrier protein (A-PCP) didomain for loading the starter unit to assemble the side chain of an Adda residue. However, the catalytic mechanism remains unclear. Here, the 2.45 Å resolution crystal structure of the McyG A-PCP didomain complexed with the catalytic intermediate L-phenylalanyl-adenylate (L-Phe-AMP) is reported. Each asymmetric unit contains two protein molecules, one of which consists of the A-PCP didomain and the other of which comprises only the A domain. Structural analyses suggest that Val227 is likely to be critical for the selection of hydrophobic substrates. Moreover, two distinct interfaces demonstrating variable crosstalk between the PCP domain and the A domain were observed. A catalytic cycle for the adenylation and peptide transfer of the A-PCP didomain is proposed.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, People's Republic of China.