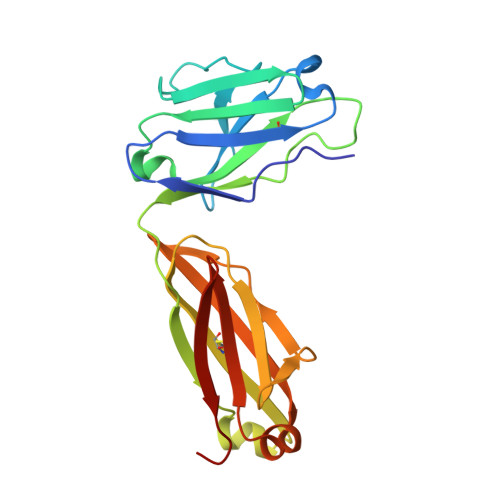
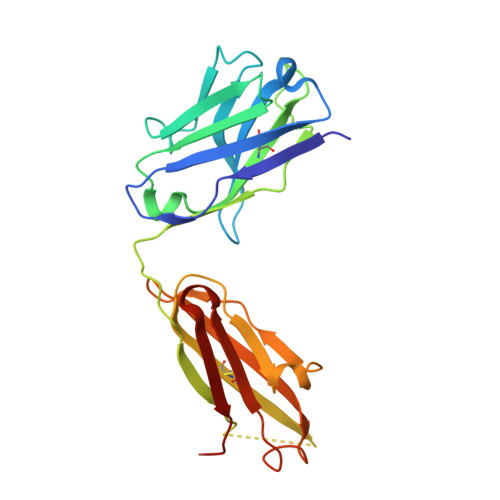
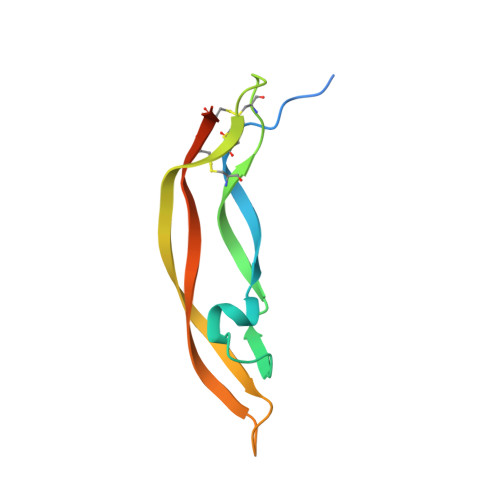
Characterization of Binding Mode of Action of a Blocking Anti-Platelet-Derived Growth Factor (PDGF)-B Monoclonal Antibody, MOR8457, Reveals Conformational Flexibility and Avidity Needed for PDGF-BB To Bind PDGF Receptor-beta.
Kuai, J., Mosyak, L., Brooks, J., Cain, M., Carven, G.J., Ogawa, S., Ishino, T., Tam, M., Lavallie, E.R., Yang, Z., Ponsel, D., Rauchenberger, R., Arch, R., Pullen, N.(2015) Biochemistry 54: 1918-1929
- PubMed: 25707433
- DOI: https://doi.org/10.1021/bi5015425
- Primary Citation of Related Structures:
4QCI - PubMed Abstract:
Platelet derived growth factor-BB (PDGF-BB) is an important mitogen and cell survival factor during development. PDGF-BB binds PDGF receptor-β (PDGFRβ) to trigger receptor dimerization and tyrosine kinase activation. We present the pharmacological and biophysical characterization of a blocking PDGF-BB monoclonal antibody, MOR8457, and contrast this to PDGFRβ. MOR8457 binds to PDGF-BB with high affinity and selectivity, and prevents PDGF-BB induced cell proliferation competitively and with high potency. The structural characterization of the MOR8457-PDGF-BB complex indicates that MOR8457 binds with a 2:1 stoichiometry, but that binding of a single MOR8457 moiety is sufficient to prevent binding to PDGFRβ. Comparison of the MOR8457-PDGF-BB structure with that of the PDGFRβ-PDGF-BB complex suggested the potential reason for this was a substantial bending and twisting of PDGF-BB in the MOR8457 structure, relative to the structures of PDGF-BB alone, bound to a PDGF-BB aptamer or PDGFRβ, which makes it nonpermissive for PDGFRβ binding. These biochemical and structural data offer insights into the permissive structure of PDGF-BB needed for agonism as well as strategies for developing specific PDGF ligand antagonists.
Organizational Affiliation:
‡Scholar Rock LLC, 300 Technology Square, Cambridge, Massachusetts 02142, United States.