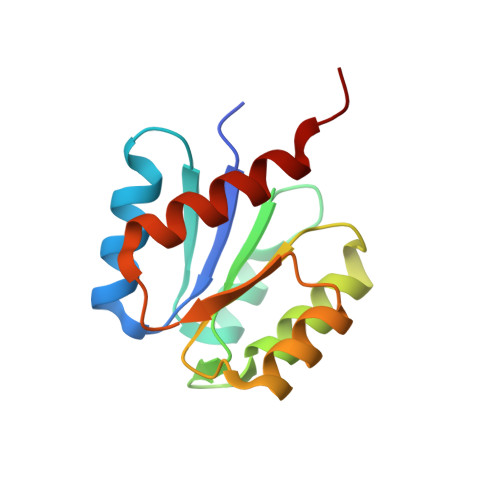
HemR is an OmpR/PhoB-like response regulator from Leptospira, which simultaneously effects transcriptional activation and repression of key haem metabolism genes.
Morero, N.R., Botti, H., Nitta, K.R., Carrion, F., Obal, G., Picardeau, M., Buschiazzo, A.(2014) Mol Microbiol 94: 340-352
- PubMed: 25145397
- DOI: https://doi.org/10.1111/mmi.12763
- Primary Citation of Related Structures:
4Q7E - PubMed Abstract:
Several Leptospira species cause leptospirosis, the most extended zoonosis worldwide. In bacteria, two-component systems constitute key signalling pathways, some of which are involved in pathogenesis. The physiological roles of two-component systems in Leptospira are largely unknown, despite identifying several dozens within their genomes. Biochemical confirmation of an operative phosphorelaying two-component system has been obtained so far only for the Hklep/Rrlep pair. It is known that hklep/rrlep knockout strains of Leptospira biflexa result in haem auxotrophy, although their de novo biosynthesis machinery remains fully functional. Haem is essential for Leptospira, but information about Hklep/Rrlep effector function(s) and target(s) is still lacking. We are now reporting a thorough molecular characterization of this system, which we rename HemK/HemR. The DNA HemR-binding motif was determined, and found within the genomes of saprophyte and pathogenic Leptospira. In this way, putative HemR-regulated genes were pinpointed, including haem catabolism-related (hmuO - haem oxygenase) and biosynthesis-related (the hemA/C/D/B/L/E/N/G operon). Specific HemR binding to these two promoters was quantified, and a dual function was observed in vivo, inversely repressing the hmuO, while activating the hemA operon transcription. The crystal structure of HemR receiver domain was determined, leading to a mechanistic model for its dual regulatory role.
Organizational Affiliation:
Institut Pasteur de Montevideo, Unit of Protein Crystallography, 2020 Mataojo, 11400, Montevideo, Uruguay.