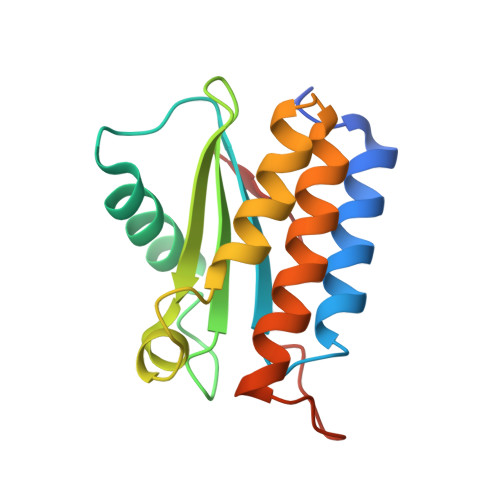
Solution and crystal structure of BA42, a protein from the Antarctic bacterium Bizionia argentinensis comprised of a stand-alone TPM domain.
Aran, M., Smal, C., Pellizza, L., Gallo, M., Otero, L.H., Klinke, S., Goldbaum, F.A., Ithurralde, E.R., Bercovich, A., Mac Cormack, W.P., Turjanski, A.G., Cicero, D.O.(2014) Proteins 82: 3062-3078
- PubMed: 25116514
- DOI: https://doi.org/10.1002/prot.24667
- Primary Citation of Related Structures:
2MPB, 4OA3 - PubMed Abstract:
The structure of the BA42 protein belonging to the Antarctic flavobacterium Bizionia argentinensis was determined by nuclear magnetic resonance and X-ray crystallography. This is the first structure of a member of the PF04536 family comprised of a stand-alone TPM domain. The structure reveals a new topological variant of the four β-strands constituting the central β-sheet of the αβα architecture and a double metal binding site stabilizing a pair of crossing loops, not observed in previous structures of proteins belonging to this family. BA42 shows differences in structure and dynamics in the presence or absence of bound metals. The affinity for divalent metal ions is close to that observed in proteins that modulate their activity as a function of metal concentration, anticipating a possible role for BA42.
Organizational Affiliation:
Fundación Instituto Leloir, IIBBA-CONICET, Patricias Argentinas 435 (C1405BWE), Buenos Aires, Argentina.