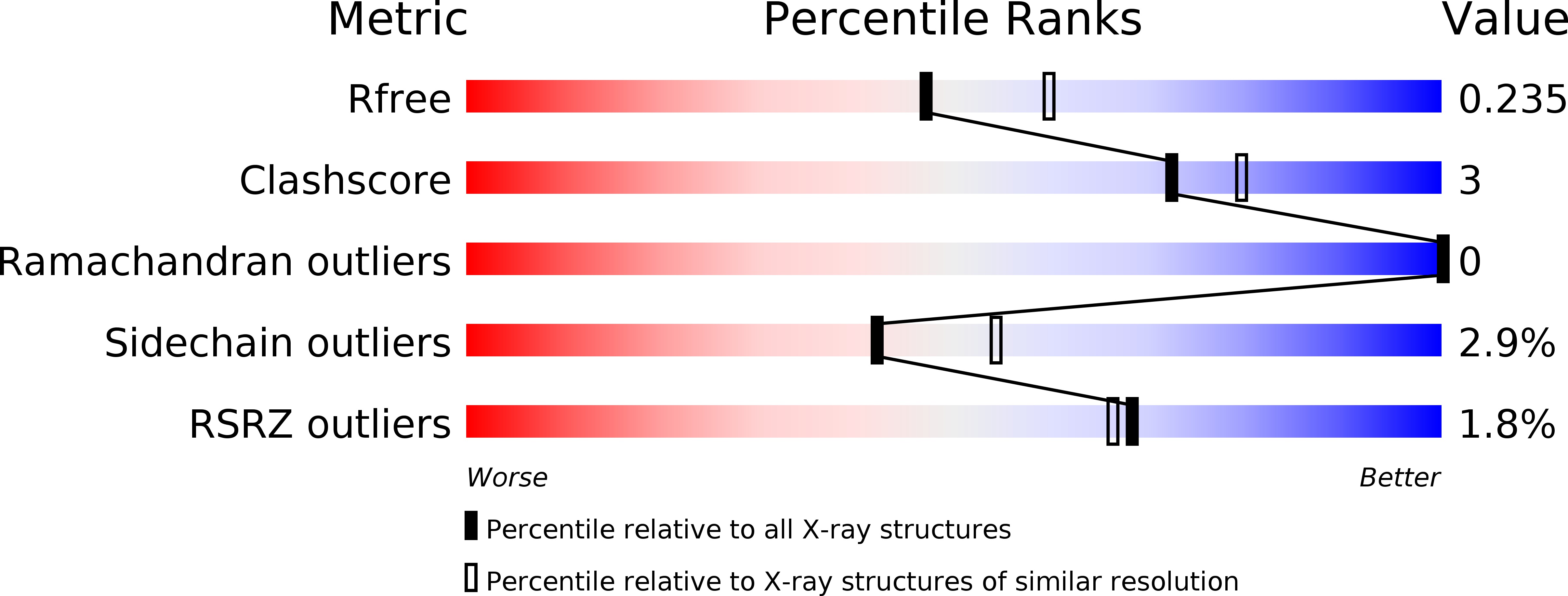
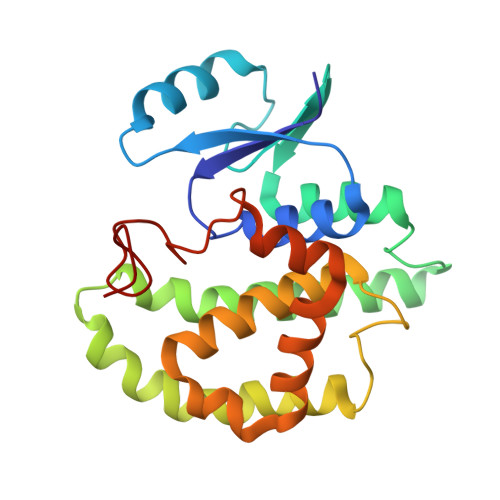
Crystal structures of 26kDa Clonorchis sinensis glutathione S-transferase reveal zinc binding and putative metal binding.
Han, Y.H., Hong, S.J., Cheong, H.K., Chung, Y.J.(2013) Biochem Biophys Res Commun 438: 457-461
- PubMed: 23916611
- DOI: https://doi.org/10.1016/j.bbrc.2013.07.102
- Primary Citation of Related Structures:
4L5L, 4L5O - PubMed Abstract:
The crystal structures of CsGST in two different space groups revealed that Asp26 and His79 coordinate a zinc ion. In one space group, His46 of an adjacent molecule participates in the coordination within 2.0Å. In the other space group, Asp26, His79 and a water molecule coordinate a zinc ion. The CsGST-D26H structure showed that four histidine residues - His26 and His79 from one molecule and the same residues from a symmetry-related neighboring molecule - coordinate a zinc ion. The coordinated zinc ions are located between two molecules and mediate molecular contacts within the crystal.
Organizational Affiliation:
Division of Magnetic Resonance Research, Korea Basic Science Institute (KBSI), Ochang, Chungbuk 363-883, Republic of Korea.