Structure of a family 3a carbohydrate-binding module from the cellulosomal scaffoldin CipA of Clostridium thermocellum with flanking linkers: implications for cellulosome structure.
Yaniv, O., Morag, E., Borovok, I., Bayer, E.A., Lamed, R., Frolow, F., Shimon, L.J.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 733-737
- PubMed: 23832198
- DOI: https://doi.org/10.1107/S174430911301614X
- Primary Citation of Related Structures:
4JO5 - PubMed Abstract:
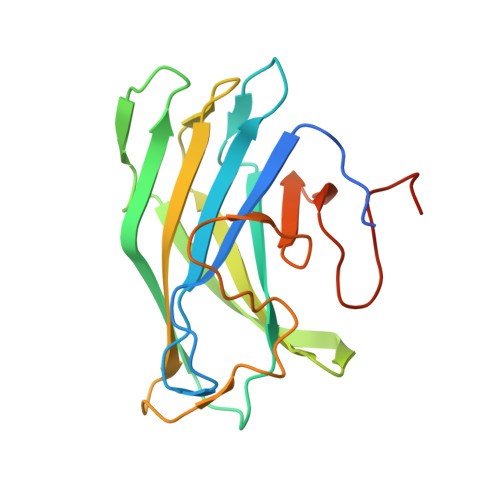
The cellulosome of the cellulolytic bacterium Clostridium thermocellum has a structural multi-modular protein called CipA (cellulosome-integrating protein A) that includes nine enzyme-binding cohesin modules and a family 3 cellulose-binding module (CBM3a). In the CipA protein, the CBM3a module is located between the second and third cohesin modules and is connected to them via proline/threonine-rich linkers. The structure of CBM3a with portions of the C- and N-terminal flanking linker regions, CBM3a-L, has been determined to a resolution of 1.98 Å. The structure is a β-sandwich with a structural Ca(2+) ion. The structure is consistent with the previously determined CipA CBM structure; however, the structured linker regions provide a deeper insight into the overall cellulosome structure and assembly.
Organizational Affiliation:
Department of Molecular Microbiology and Biotechnology, Tel Aviv University, 69978 Tel Aviv, Israel.