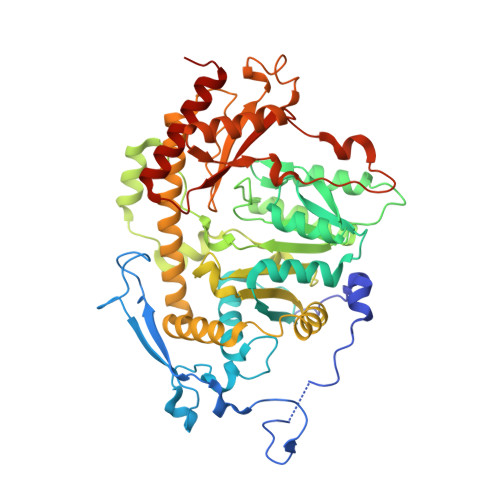
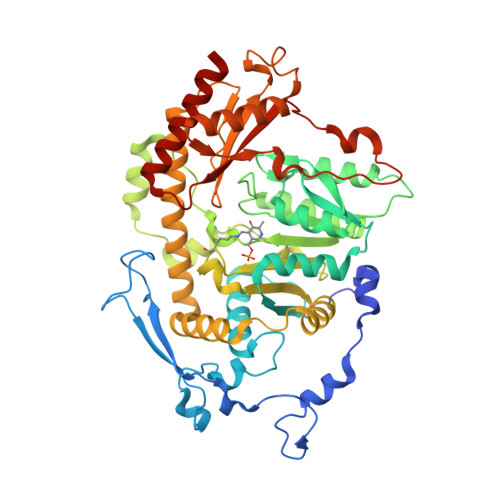
Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase.
Bulfer, S.L., Brunzelle, J.S., Trievel, R.C.(2013) Protein Sci 22: 1417-1424
- PubMed: 23893908
- DOI: https://doi.org/10.1002/pro.2315
- Primary Citation of Related Structures:
4JE5 - PubMed Abstract:
α-Aminoadipate aminotransferase (AAA-AT) catalyzes the amination of 2-oxoadipate to α-aminoadipate in the fourth step of the α-aminoadipate pathway of lysine biosynthesis in fungi. The aromatic aminotransferase Aro8 has recently been identified as an AAA-AT in Saccharomyces cerevisiae. This enzyme displays broad substrate selectivity, utilizing several amino acids and 2-oxo acids as substrates. Here we report the 1.91Å resolution crystal structure of Aro8 and compare it to AAA-AT LysN from Thermus thermophilus and human kynurenine aminotransferase II. Inspection of the active site of Aro8 reveals asymmetric cofactor binding with lysine-pyridoxal-5-phosphate bound within the active site of one subunit in the Aro8 homodimer and pyridoxamine phosphate and a HEPES molecule bound to the other subunit. The HEPES buffer molecule binds within the substrate-binding site of Aro8, yielding insights into the mechanism by which it recognizes multiple substrates and how this recognition differs from other AAA-AT/kynurenine aminotransferases.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor, Michigan, 48109.