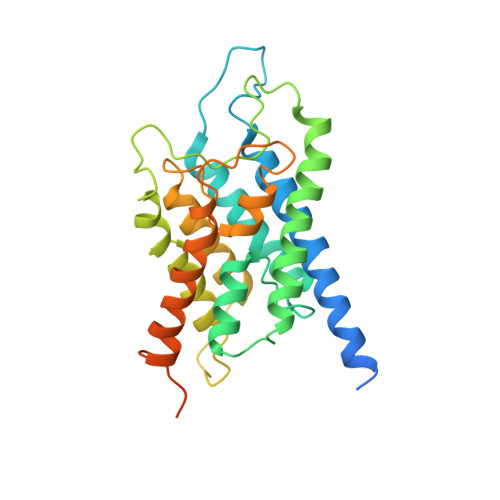
Crystallization and Preliminary Crystallographic Analysis of Human Aquaporin 1 at a Resolution of 3.28 A.
Ruiz Carrillo, D., To Yiu Ying, J., Darwis, D., Soon, C.H., Cornvik, T., Torres, J., Lescar, J.(2014) Acta Crystallogr Sect F Struct Biol Cryst Commun 70: 1657
- PubMed: 25484221
- DOI: https://doi.org/10.1107/S2053230X14024558
- Primary Citation of Related Structures:
4CSK - PubMed Abstract:
Aquaporin water channels (AQPs) are found in almost every organism from humans to bacteria. In humans, 13 classes of AQPs control water and glycerol homeostasis. Knockout studies have suggested that modulating the activity of AQPs could be beneficial for the treatment of several pathologies. In particular, aquaporin 1 is a key factor in cell migration and angiogenesis, and constitutes a possible target for anticancer compounds and also for the treatment of glaucoma. Here, a preliminary crystallographic analysis at 3.28 Å resolution of crystals of human aquaporin 1 (hAQP1) obtained from protein expressed in Sf9 insect cells is reported. The crystals belonged to the tetragonal space group I422, with unit-cell parameters a = b = 89.28, c = 174.9 Å, and contained one monomer per asymmetric unit. The hAQP1 biological tetramer is generated via the crystallographic fourfold axis. This work extends previous electron crystallographic studies that used material extracted from human red blood cells, in which the resolution was limited to approximately 3.8 Å. It will inform efforts to improve lattice contacts and the diffraction limit for the future structure-based discovery of specific hAQP1 inhibitors.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 61 Biopolis Drive, Singapore 138673, Singapore.