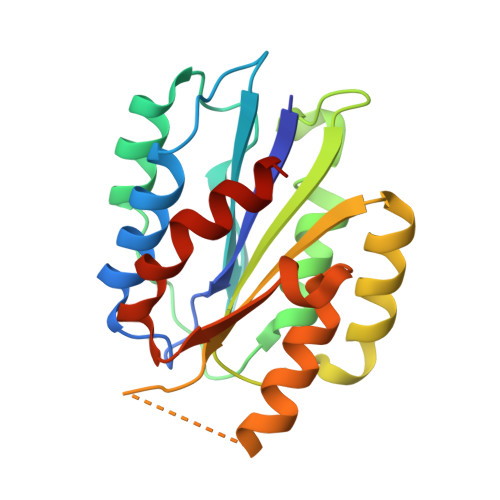
Structure of Collagen Receptor Integrin Aplha1I Domain Carrying the Activating Mutation E317A.
Lahti, M., Bligt, E., Niskanen, H., Parkash, V., Brandt, A.M., Jokinen, J., Patrikainen, P., Kapyla, J., Heino, J., Salminen, T.A.(2011) J Biol Chem 286: 43343
- PubMed: 22030389
- DOI: https://doi.org/10.1074/jbc.M111.261909
- Primary Citation of Related Structures:
4A0Q - PubMed Abstract:
We have analyzed the structure and function of the integrin α(1)I domain harboring a gain-of-function mutation E317A. To promote protein crystallization, a double variant with an additional C139S mutation was used. In cell adhesion assays, the E317A mutation promoted binding to collagen. Similarly, the double mutation C139S/E317A increased adhesion compared with C139S alone. Furthermore, soluble α(1)I C139S/E317A was a higher avidity collagen binder than α(1)I C139S, indicating that the double variant represents an activated form. The crystal structure of the activated variant of α(1)I was solved at 1.9 Å resolution. The E317A mutation results in the unwinding of the αC helix, but the metal ion has moved toward loop 1, instead of loop 2 in the open α(2)I. Furthermore, unlike in the closed αI domains, the metal ion is pentacoordinated and, thus, prepared for ligand binding. Helix 7, which has moved downward in the open α(2)I structure, has not changed its position in the activated α(1)I variant. During the integrin activation, Glu(335) on helix 7 binds to the metal ion at the metal ion-dependent adhesion site (MIDAS) of the β(1) subunit. Interestingly, in our cell adhesion assays E317A could activate collagen binding even after mutating Glu(335). This indicates that the stabilization of helix 7 into its downward position is not required if the α(1) MIDAS is already open. To conclude, the activated α(1)I domain represents a novel conformation of the αI domain, mimicking the structural state where the Arg(287)-Glu(317) ion pair has just broken during the integrin activation.
Organizational Affiliation:
Department of Biochemistry and Food Chemistry, University of Turku, Turku FI-20014, Finland.